Translation directed by hepatitis A virus IRES in the absence of active eIF4F complex and eIF2
- PMID: 23272212
- PMCID: PMC3525551
- DOI: 10.1371/journal.pone.0052065
Translation directed by hepatitis A virus IRES in the absence of active eIF4F complex and eIF2
Abstract
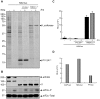
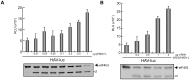
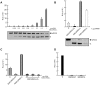
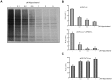
Translation directed by several picornavirus IRES elements can usually take place after cleavage of eIF4G by picornavirus proteases 2A(pro) or L(pro). The hepatitis A virus (HAV) IRES is thought to be an exception to this rule because it requires intact eIF4F complex for translation. In line with previous results we report that poliovirus (PV) 2A(pro) strongly blocks protein synthesis directed by HAV IRES. However, in contrast to previous findings we now demonstrate that eIF4G cleavage by foot-and-mouth disease virus (FMDV) L(pro) strongly stimulates HAV IRES-driven translation. Thus, this is the first observation that 2A(pro) and L(pro) exhibit opposite effects to what was previously thought to be the case in HAV IRES. This effect has been observed both in hamster BHK and human hepatoma Huh7 cells. In addition, this stimulation of translation is also observed in cell free systems after addition of purified L(pro). Notably, in presence of this FMDV protease, translation directed by HAV IRES takes place when eIF2α has been inactivated by phosphorylation. Our present findings clearly demonstrate that protein synthesis directed by HAV IRES can occur when eIF4G has been cleaved and after inactivation of eIF2. Therefore, translation directed by HAV IRES without intact eIF4G and active eIF2 is similar to that observed with other picornavirus IRESs.
Conflict of interest statement
Figures


Similar articles
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
-
L protease from foot and mouth disease virus confers eIF2-independent translation for mRNAs bearing picornavirus IRES.FEBS Lett. 2014 Nov 3;588(21):4053-9. doi: 10.1016/j.febslet.2014.09.030. Epub 2014 Sep 28. FEBS Lett. 2014. PMID: 25268112
-
Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation.Proc Natl Acad Sci U S A. 2017 Sep 5;114(36):9611-9616. doi: 10.1073/pnas.1704390114. Epub 2017 Aug 21. Proc Natl Acad Sci U S A. 2017. PMID: 28827335 Free PMC article.
-
Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F.J Virol. 2001 Sep;75(17):7864-71. doi: 10.1128/jvi.75.17.7864-7871.2001. J Virol. 2001. PMID: 11483730 Free PMC article.
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
Cited by
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
-
Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding.Proc Natl Acad Sci U S A. 2017 Jun 13;114(24):6310-6315. doi: 10.1073/pnas.1610417114. Epub 2017 May 30. Proc Natl Acad Sci U S A. 2017. PMID: 28559344 Free PMC article.
-
Response of Three Different Viruses to Interferon Priming and Dithiothreitol Treatment of Avian Cells.J Virol. 2016 Aug 26;90(18):8328-40. doi: 10.1128/JVI.01175-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27440902 Free PMC article.
-
RNA-binding proteins impacting on internal initiation of translation.Int J Mol Sci. 2013 Nov 1;14(11):21705-26. doi: 10.3390/ijms141121705. Int J Mol Sci. 2013. PMID: 24189219 Free PMC article. Review.
-
The Regulation of Translation in Alphavirus-Infected Cells.Viruses. 2018 Feb 8;10(2):70. doi: 10.3390/v10020070. Viruses. 2018. PMID: 29419763 Free PMC article. Review.
References
-
- Belsham GJ (2009) Divergent picornavirus IRES elements. Virus research 139: 183–192. - PubMed
-
- Niepmann M (2009) Internal translation initiation of picornaviruses and hepatitis C virus. Biochimica et biophysica acta 1789: 529–541. - PubMed
-
- Borman AM, Kean KM (1997) Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237: 129–136. - PubMed

