Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection
- PMID: 23273270
- PMCID: PMC3552703
- DOI: 10.1186/1758-907X-3-9
Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection
Abstract
Background: High throughput DNA sequencing technology has enabled quantification of all the RNAs in a cell or tissue, a method widely known as RNA sequencing (RNA-Seq). However, non-coding RNAs such as rRNA are highly abundant and can consume >70% of sequencing reads. A common approach is to extract only polyadenylated mRNA; however, such approaches are blind to RNAs with short or no poly(A) tails, leading to an incomplete view of the transcriptome. Another challenge of preparing RNA-Seq libraries is to preserve the strand information of the RNAs.
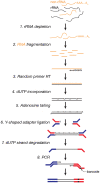
Design: Here, we describe a procedure for preparing RNA-Seq libraries from 1 to 4 μg total RNA without poly(A) selection. Our method combines the deoxyuridine triphosphate (dUTP)/uracil-DNA glycosylase (UDG) strategy to achieve strand specificity with AMPure XP magnetic beads to perform size selection. Together, these steps eliminate gel purification, allowing a library to be made in less than two days. We barcode each library during the final PCR amplification step, allowing several samples to be sequenced in a single lane without sacrificing read length. Libraries prepared using this protocol are compatible with Illumina GAII, GAIIx and HiSeq 2000 platforms.
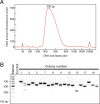
Discussion: The RNA-Seq protocol described here yields strand-specific transcriptome libraries without poly(A) selection, which provide approximately 90% mappable sequences. Typically, more than 85% of mapped reads correspond to protein-coding genes and only 6% derive from non-coding RNAs. The protocol has been used to measure RNA transcript identity and abundance in tissues from flies, mice, rats, chickens, and frogs, demonstrating its general applicability.
Figures
References
-
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2009;11:31–46. - PubMed
-
- Huang R, Jaritz M, Guenzl P, Vlatkovic I, Sommer A, Tamir IM, Marks H, Klampfl T, Kralovics R, Stunnenberg HG. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS One. 2011;6:e27288. doi: 10.1371/journal.pone.0027288. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources