The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: implications for transcription start-site selection
- PMID: 23274143
- PMCID: PMC3783996
- DOI: 10.1016/j.jmb.2012.12.015
The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: implications for transcription start-site selection
Abstract
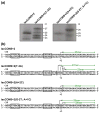
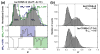
Bacterial transcription is initiated after RNA polymerase (RNAP) binds to promoter DNA, melts ~14 bp around the transcription start site and forms a single-stranded "transcription bubble" within a catalytically active RNAP-DNA open complex (RP(o)). There is significant flexibility in the transcription start site, which causes variable spacing between the promoter elements and the start site; this in turn causes differences in the length and sequence at the 5' end of RNA transcripts and can be important for gene regulation. The start-site variability also implies the presence of some flexibility in the positioning of the DNA relative to the RNAP active site in RP(o). The flexibility may occur in the positioning of the transcription bubble prior to RNA synthesis and may reflect bubble expansion ("scrunching") or bubble contraction ("unscrunching"). Here, we assess the presence of dynamic flexibility in RP(o) with single-molecule FRET (Förster resonance energy transfer). We obtain experimental evidence for dynamic flexibility in RP(o) using different FRET rulers and labeling positions. An analysis of FRET distributions of RP(o) using burst variance analysis reveals conformational fluctuations in RP(o) in the millisecond timescale. Further experiments using subsets of nucleotides and DNA mutations allowed us to reprogram the transcription start sites, in a way that can be described by repositioning of the single-stranded transcription bubble relative to the RNAP active site within RP(o). Our study marks the first experimental observation of conformational dynamics in the transcription bubble of RP(o) and indicates that DNA dynamics within the bubble affect the search for transcription start sites.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Carpousis AJ, Stefano JE, Gralla JD. 5′ Nucleotide heterogeneity and altered initiation of transcription at mutant lac promoters. J Mol Biol. 1982;157:619–633. - PubMed
-
- Minkley EG, Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973;77:255–277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

