Interplay between viruses and host mRNA degradation
- PMID: 23274304
- PMCID: PMC3632658
- DOI: 10.1016/j.bbagrm.2012.12.003
Interplay between viruses and host mRNA degradation
Abstract
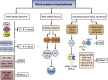
Messenger RNA degradation is a fundamental cellular process that plays a critical role in regulating gene expression by controlling both the quality and the abundance of mRNAs in cells. Naturally, viruses must successfully interface with the robust cellular RNA degradation machinery to achieve an optimal balance between viral and cellular gene expression and establish a productive infection in the host. In the past several years, studies have discovered many elegant strategies that viruses have evolved to circumvent the cellular RNA degradation machinery, ranging from disarming the RNA decay pathways and co-opting the factors governing cellular mRNA stability to promoting host mRNA degradation that facilitates selective viral gene expression and alters the dynamics of host-pathogen interaction. This review summarizes the current knowledge of the multifaceted interaction between viruses and cellular mRNA degradation machinery to provide an insight into the regulatory mechanisms that influence gene expression in viral infections. This article is part of a Special Issue entitled: RNA Decay mechanisms.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Inhibition and avoidance of mRNA degradation by RNA viruses.Curr Opin Microbiol. 2012 Aug;15(4):500-5. doi: 10.1016/j.mib.2012.04.009. Epub 2012 May 23. Curr Opin Microbiol. 2012. PMID: 22626865 Free PMC article. Review.
-
Viruses and the cellular RNA decay machinery.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):47-59. doi: 10.1002/wrna.3. Epub 2010 May 6. Wiley Interdiscip Rev RNA. 2010. PMID: 21956906 Free PMC article. Review.
-
C19ORF66 Broadly Escapes Virus-Induced Endonuclease Cleavage and Restricts Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2019 May 29;93(12):e00373-19. doi: 10.1128/JVI.00373-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944177 Free PMC article.
-
Fated for decay: RNA elements targeted by viral endonucleases.Semin Cell Dev Biol. 2021 Mar;111:119-125. doi: 10.1016/j.semcdb.2020.05.010. Epub 2020 Jun 7. Semin Cell Dev Biol. 2021. PMID: 32522410 Free PMC article. Review.
-
Viral Evasion and Manipulation of Host RNA Quality Control Pathways.J Virol. 2016 Jul 27;90(16):7010-7018. doi: 10.1128/JVI.00607-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27226372 Free PMC article. Review.
Cited by
-
The Lsm1-7-Pat1 complex promotes viral RNA translation and replication by differential mechanisms.RNA. 2015 Aug;21(8):1469-79. doi: 10.1261/rna.052209.115. Epub 2015 Jun 19. RNA. 2015. PMID: 26092942 Free PMC article.
-
Abscisic Acid Connects Phytohormone Signaling with RNA Metabolic Pathways and Promotes an Antiviral Response that Is Evaded by a Self-Controlled RNA Virus.Plant Commun. 2020 Sep 14;1(5):100099. doi: 10.1016/j.xplc.2020.100099. Epub 2020 Jul 7. Plant Commun. 2020. PMID: 32984814 Free PMC article.
-
Structures and Functions of the 3' Untranslated Regions of Positive-Sense Single-Stranded RNA Viruses Infecting Humans and Animals.Front Cell Infect Microbiol. 2020 Aug 27;10:453. doi: 10.3389/fcimb.2020.00453. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32974223 Free PMC article. Review.
-
Fast and efficient neural conversion of human hematopoietic cells.Stem Cell Reports. 2014 Dec 9;3(6):1118-31. doi: 10.1016/j.stemcr.2014.10.008. Epub 2014 Nov 13. Stem Cell Reports. 2014. PMID: 25458894 Free PMC article.
-
Truncation of C-terminal 20 amino acids in PA-X contributes to adaptation of swine influenza virus in pigs.Sci Rep. 2016 Feb 25;6:21845. doi: 10.1038/srep21845. Sci Rep. 2016. PMID: 26912401 Free PMC article.
References
-
- Komili S., Silver P.A. Coupling and coordination in gene expression processes: a systems biology view. Nat. Rev. Genet. 2008;9:38–48. - PubMed
-
- Sachs A.B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. - PubMed
-
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. - PubMed
-
- Behm-Ansmant I., Kashima I., Rehwinkel J., Sauliere J., Wittkopp N., Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

