Function of cytochrome P450 enzymes RosC and RosD in the biosynthesis of rosamicin macrolide antibiotic produced by Micromonospora rosaria
- PMID: 23274670
- PMCID: PMC3591866
- DOI: 10.1128/AAC.02092-12
Function of cytochrome P450 enzymes RosC and RosD in the biosynthesis of rosamicin macrolide antibiotic produced by Micromonospora rosaria
Abstract
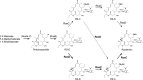
The cytochrome P450 enzyme-encoding genes rosC and rosD were cloned from the rosamicin biosynthetic gene cluster of Micromonospora rosaria IFO13697. The functions of RosC and RosD were demonstrated by gene disruption and complementation with M. rosaria and bioconversion of rosamicin biosynthetic intermediates with Escherichia coli expressing RosC and RosD. It is proposed that M. rosaria IFO13697 has two pathway branches that lead from the first desosaminyl rosamicin intermediate, 20-deoxo-20-dihydro-12,13-deepoxyrosamicin, to rosamicin.
Figures

Similar articles
-
A new mycinosyl rosamicin derivative produced by an engineered Micromonospora rosaria mutant with a cytochrome P450 gene disruption introducing the D-mycinose biosynthetic gene.J Ind Microbiol Biotechnol. 2014 Sep;41(9):1451-6. doi: 10.1007/s10295-014-1488-2. Epub 2014 Jul 22. J Ind Microbiol Biotechnol. 2014. PMID: 25047182
-
Cytochrome P450 enzyme RosC catalyzes a multistep oxidation reaction to form the non-active compound 20-carboxyrosamicin.FEMS Microbiol Lett. 2017 Jul 3;364(12). doi: 10.1093/femsle/fnx110. FEMS Microbiol Lett. 2017. PMID: 28582504
-
Artificial control of the multistep oxidation reactions catalyzed by the cytochrome P450 enzyme RosC.Appl Microbiol Biotechnol. 2020 Apr;104(8):3403-3415. doi: 10.1007/s00253-020-10481-7. Epub 2020 Feb 26. Appl Microbiol Biotechnol. 2020. PMID: 32103316
-
Macrolide antibiotics, drug interactions and microsomal enzymes: implications for veterinary medicine.Res Vet Sci. 1999 Jun;66(3):197-203. doi: 10.1053/rvsc.1998.0244. Res Vet Sci. 1999. PMID: 10333459 Review.
-
Application of Genetic Engineering Approaches to Improve Bacterial Metabolite Production.Curr Protein Pept Sci. 2020;21(5):488-496. doi: 10.2174/1389203721666191223145827. Curr Protein Pept Sci. 2020. PMID: 31868145 Review.
Cited by
-
Chemoenzymatic Total Synthesis and Structural Diversification of Tylactone-Based Macrolide Antibiotics through Late-Stage Polyketide Assembly, Tailoring, and C-H Functionalization.J Am Chem Soc. 2017 Jun 14;139(23):7913-7920. doi: 10.1021/jacs.7b02875. Epub 2017 Jun 5. J Am Chem Soc. 2017. PMID: 28525276 Free PMC article.
-
Post-PKS tailoring steps of the spiramycin macrolactone ring in Streptomyces ambofaciens.Antimicrob Agents Chemother. 2013 Aug;57(8):3836-42. doi: 10.1128/AAC.00512-13. Epub 2013 May 28. Antimicrob Agents Chemother. 2013. PMID: 23716060 Free PMC article.
-
A new mycinosyl rosamicin derivative produced by an engineered Micromonospora rosaria mutant with a cytochrome P450 gene disruption introducing the D-mycinose biosynthetic gene.J Ind Microbiol Biotechnol. 2014 Sep;41(9):1451-6. doi: 10.1007/s10295-014-1488-2. Epub 2014 Jul 22. J Ind Microbiol Biotechnol. 2014. PMID: 25047182
-
Targeting Reactive Carbonyls for Identifying Natural Products and Their Biosynthetic Origins.J Am Chem Soc. 2016 Nov 23;138(46):15157-15166. doi: 10.1021/jacs.6b06848. Epub 2016 Nov 14. J Am Chem Soc. 2016. PMID: 27797509 Free PMC article.
-
Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.Nat Commun. 2023 Mar 25;14(1):1669. doi: 10.1038/s41467-023-37288-0. Nat Commun. 2023. PMID: 36966128 Free PMC article.
References
-
- Puar MS, Schumacher D. 1990. Novel macrolides from Micromonospora rosaria. J. Antibiot. 43:1497–1501 - PubMed
-
- Anzai Y, Iizaka Y, Li W, Idemoto N, Tsukada S, Koike K, Kinoshita K, Kato F. 2009. Production of rosamicin derivatives in Micromonospora rosaria by introduction of d-mycinose biosynthetic gene with ϕC31-derived integration vector pSET152. J. Ind. Microbiol. Biotechnol. 36:1013–1021 - PubMed
-
- Anzai Y, Sakai A, Li W, Iizaka Y, Koike K, Kinoshita K, Kato F. 2010. Isolation and characterization of 23-O-mycinosyl-20-dihydro-rosamicin: a new rosamicin analogue derived from engineered Micromonospora rosaria. J. Antibiot. 63:325–328 - PubMed
-
- Merson-Davies LA, Cundliffe E. 1994. Analysis of five tylosin biosynthetic genes from the tyllBA region of the Streptomyces fradiae genome. Mol. Microbiol. 13:349–355 - PubMed
-
- Schell U, Haydock SF, Kaja AL, Carletti I, Lill RE, Read E, Sheehan LS, Low L, Fernandez MJ, Grolle F, McArthur HA, Sheridan RM, Leadlay PF, Wilkinson B, Gaisser S. 2008. Engineered biosynthesis of hybrid macrolide polyketides containing d-angolosamine and d-mycaminose moieties. Org. Biomol. Chem. 6:3315–3327 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

