The unique structure of Haemophilus influenzae protein E reveals multiple binding sites for host factors
- PMID: 23275089
- PMCID: PMC3584867
- DOI: 10.1128/IAI.01111-12
The unique structure of Haemophilus influenzae protein E reveals multiple binding sites for host factors
Abstract
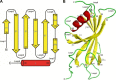
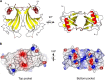
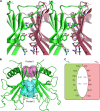
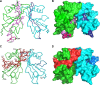
Haemophilus influenzae protein E (PE) is a multifunctional adhesin involved in direct interactions with lung epithelial cells and host proteins, including plasminogen and the extracellular matrix proteins vitronectin and laminin. We recently crystallized PE and successfully collected X-ray diffraction data at 1.8 Å. Here, we solved the structure of a recombinant version of PE and analyzed different functional regions. It is a dimer in solution and in the asymmetric unit of the crystals. The dimer has a structure that resembles a flattened β-barrel. It is, however, not a true β-barrel, as there are differences in both the hydrogen-bonding pattern and the shape. Each monomer consisted of a 6-stranded antiparallel β-sheet with a rigid α-helix at the C terminus tethered to the concave side of the sheet by a disulfide bridge. The laminin/plasminogen binding region (residues 41 to 68) is exposed, while the vitronectin binding region (residues 84 to 108) is partially accessible in the dimer. The dimerized PE explains the simultaneous interaction with laminin and vitronectin. In addition, we found this unique adhesin to be present in many bacterial genera of the family Pasteurellaceae and also orthologues in other, unrelated species (Enterobacter cloacae and Listeria monocytogenes). Peptides corresponding to the surface-exposed regions PE 24 to 37, PE 74 to 89, and PE 134 to 156 were immunogenic in the mouse. Importantly, these peptide-based antibodies also recognized PE at the bacterial surface. Taken together, our detailed structure of PE explains how this important virulence factor of H. influenzae simultaneously interacts with host vitronectin, laminin, or plasminogen, promoting bacterial pathogenesis.
Figures




Similar articles
-
Crystallization and X-ray diffraction analysis of a novel surface-adhesin protein: protein E from Haemophilus influenzae.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Feb 1;68(Pt 2):222-6. doi: 10.1107/S1744309111055503. Epub 2012 Jan 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22298005 Free PMC article.
-
Haemophilus influenzae uses the surface protein E to acquire human plasminogen and to evade innate immunity.J Immunol. 2012 Jan 1;188(1):379-85. doi: 10.4049/jimmunol.1101927. Epub 2011 Nov 28. J Immunol. 2012. PMID: 22124123
-
Haemophilus influenzae stores and distributes hemin by using protein E.Int J Med Microbiol. 2014 Jul;304(5-6):662-8. doi: 10.1016/j.ijmm.2014.04.015. Epub 2014 May 9. Int J Med Microbiol. 2014. PMID: 24863527
-
Haemophilus influenzae protein E binds to the extracellular matrix by concurrently interacting with laminin and vitronectin.J Infect Dis. 2011 Oct 1;204(7):1065-74. doi: 10.1093/infdis/jir459. J Infect Dis. 2011. PMID: 21881122
-
In vitro selection of RNA aptamers directed against protein E: a Haemophilus influenzae adhesin.Mol Biotechnol. 2014 Aug;56(8):714-25. doi: 10.1007/s12033-014-9749-x. Mol Biotechnol. 2014. PMID: 24682699
Cited by
-
Antibodies to the HMW1/HMW2 and Hia adhesins of nontypeable haemophilus influenzae mediate broad-based opsonophagocytic killing of homologous and heterologous strains.Clin Vaccine Immunol. 2014 May;21(5):613-21. doi: 10.1128/CVI.00772-13. Epub 2014 Feb 26. Clin Vaccine Immunol. 2014. PMID: 24574538 Free PMC article.
-
The Laminin Interactome: A Multifactorial Laminin-Binding Strategy by Nontypeable Haemophilus influenzae for Effective Adherence and Colonization.J Infect Dis. 2019 Aug 9;220(6):1049-1060. doi: 10.1093/infdis/jiz217. J Infect Dis. 2019. PMID: 31034569 Free PMC article.
-
How bacteria hack the matrix and dodge the bullets of immunity.Eur Respir Rev. 2018 Jun 27;27(148):180018. doi: 10.1183/16000617.0018-2018. Print 2018 Jun 30. Eur Respir Rev. 2018. PMID: 29950304 Free PMC article. Review.
-
Anaplasma phagocytophilum surface protein AipA mediates invasion of mammalian host cells.Cell Microbiol. 2014 Aug;16(8):1133-45. doi: 10.1111/cmi.12286. Epub 2014 Apr 3. Cell Microbiol. 2014. PMID: 24612118 Free PMC article.
-
Identification of antigenic proteins of the nosocomial pathogen Klebsiella pneumoniae.PLoS One. 2014 Oct 21;9(10):e110703. doi: 10.1371/journal.pone.0110703. eCollection 2014. PLoS One. 2014. PMID: 25333280 Free PMC article.
References
-
- Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr. Infect. Dis. J. 28:43–48 - PubMed
-
- van Wessel K, Rodenburg GD, Veenhoven RH, Spanjaard L, van der Ende A, Sanders EA. 2011. Nontypeable Haemophilus influenzae invasive disease in The Netherlands: a retrospective surveillance study 2001–2008. Clin. Infect. Dis. 53:e1–e7 doi:10.1093/cid/cir268. - DOI - PubMed
-
- Resman F, Ristovski M, Ahl J, Forsgren A, Gilsdorf JR, Jasir A, Kaijser B, Kronvall G, Riesbeck K. 2011. Invasive disease caused by Haemophilus influenzae in Sweden 1997–2009; evidence of increasing incidence and clinical burden of non-type b strains. Clin. Microbiol. Infect. 17:1638–1645 - PubMed
-
- Grandi G. 2001. Antibacterial vaccine design using genomics and proteomics. Trends Biotechnol. 19:181–188 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

