Synthesis and decoding of selenocysteine and human health
- PMID: 23275319
- PMCID: PMC3541580
- DOI: 10.3325/cmj.2012.53.535
Synthesis and decoding of selenocysteine and human health
Abstract
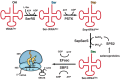
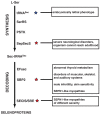
Selenocysteine, the 21st amino acid, has been found in 25 human selenoproteins and selenoenzymes important for fundamental cellular processes ranging from selenium homeostasis maintenance to the regulation of the overall metabolic rate. In all organisms that contain selenocysteine, both the synthesis of selenocysteine and its incorporation into a selenoprotein requires an elaborate synthetic and translational apparatus, which does not resemble the canonical enzymatic system employed for the 20 standard amino acids. In humans, three synthetic enzymes, a specialized elongation factor, an accessory protein factor, two catabolic enzymes, a tRNA, and a stem-loop structure in the selenoprotein mRNA are critical for ensuring that only selenocysteine is attached to selenocysteine tRNA and that only selenocysteine is inserted into the nascent polypeptide in response to a context-dependent UGA codon. The abnormal selenium homeostasis and mutations in selenoprotein genes have been causatively linked to a variety of human diseases, which, in turn, sparked a renewed interest in utilizing selenium as the dietary supplement to either prevent or remedy pathologic conditions. In contrast, the importance of the components of the selenocysteine-synthetic machinery for human health is less clear. Emerging evidence suggests that enzymes responsible for selenocysteine formation and decoding the selenocysteine UGA codon, which by extension are critical for synthesis of the entire selenoproteome, are essential for the development and health of the human organism.
Figures


References
-
- Klaproth MH. Chemical examination of the Transylvanian gold ores, collection of German papers, which were read aloud at the Royal Academy of Sciences in Berlin in the years 1789-1800 [in German]. 1803;15.
-
- Berzelius JJ.Letter from Berthollet Berzelius M on two new metals[in French]Annales de Chimie et de Physique, Série 2 18187199–202.
-
- Amor AJ, Pringle P. A review of selenium as an industrial hazard. Bull Hyg (Lond) 1945;20:239–41.
-
- Painter EP. The chemistry and toxicity of selenium compounds, with special reference to the selnium problem. Chem Rev. 1941;28:179–213. doi: 10.1021/cr60090a001. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources