Origins of shared genetic variation in African cichlids
- PMID: 23275489
- PMCID: PMC3603313
- DOI: 10.1093/molbev/mss326
Origins of shared genetic variation in African cichlids
Abstract
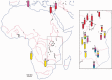
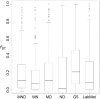
Cichlid fishes have evolved tremendous morphological and behavioral diversity in the waters of East Africa. Within each of the Great Lakes Tanganyika, Malawi, and Victoria, the phenomena of hybridization and retention of ancestral polymorphism explain allele sharing across species. Here, we explore the sharing of single nucleotide polymorphisms (SNPs) between the major East African cichlid assemblages. A set of approximately 200 genic and nongenic SNPs was ascertained in five Lake Malawi species and genotyped in a diverse collection of ~160 species from across Africa. We observed segregating polymorphism outside of the Malawi lineage for more than 50% of these loci; this holds similarly for genic versus nongenic SNPs, as well as for SNPs at putative CpG versus non-CpG sites. Bayesian and principal component analyses of genetic structure in the data demonstrate that the Lake Malawi endemic flock is not monophyletic and that river species have likely contributed significantly to Malawi genomes. Coalescent simulations support the hypothesis that river cichlids have transported polymorphism between lake assemblages. We observed strong genetic differentiation between Malawi lineages for approximately 8% of loci, with contributions from both genic and nongenic SNPs. Notably, more than half of these outlier loci between Malawi groups are polymorphic outside of the lake. Cichlid fishes have evolved diversity in Lake Malawi as new mutations combined with standing genetic variation shared across East Africa.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

