Molecular analysis of the in situ growth rates of subsurface Geobacter species
- PMID: 23275510
- PMCID: PMC3591973
- DOI: 10.1128/AEM.03263-12
Molecular analysis of the in situ growth rates of subsurface Geobacter species
Abstract
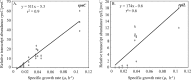
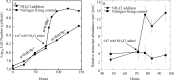
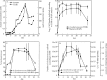
Molecular tools that can provide an estimate of the in situ growth rate of Geobacter species could improve understanding of dissimilatory metal reduction in a diversity of environments. Whole-genome microarray analyses of a subsurface isolate of Geobacter uraniireducens, grown under a variety of conditions, identified a number of genes that are differentially expressed at different specific growth rates. Expression of two genes encoding ribosomal proteins, rpsC and rplL, was further evaluated with quantitative reverse transcription-PCR (qRT-PCR) in cells with doubling times ranging from 6.56 h to 89.28 h. Transcript abundance of rpsC correlated best (r(2) = 0.90) with specific growth rates. Therefore, expression patterns of rpsC were used to estimate specific growth rates of Geobacter species during an in situ uranium bioremediation field experiment in which acetate was added to the groundwater to promote dissimilatory metal reduction. Initially, increased availability of acetate in the groundwater resulted in higher expression of Geobacter rpsC, and the increase in the number of Geobacter cells estimated with fluorescent in situ hybridization compared well with specific growth rates estimated from levels of in situ rpsC expression. However, in later phases, cell number increases were substantially lower than predicted from rpsC transcript abundance. This change coincided with a bloom of protozoa and increased attachment of Geobacter species to solid phases. These results suggest that monitoring rpsC expression may better reflect the actual rate that Geobacter species are metabolizing and growing during in situ uranium bioremediation than changes in cell abundance.
Figures

Similar articles
-
Enrichment of specific protozoan populations during in situ bioremediation of uranium-contaminated groundwater.ISME J. 2013 Jul;7(7):1286-98. doi: 10.1038/ismej.2013.20. Epub 2013 Feb 28. ISME J. 2013. PMID: 23446832 Free PMC article.
-
Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments.ISME J. 2009 Feb;3(2):216-30. doi: 10.1038/ismej.2008.89. Epub 2008 Oct 9. ISME J. 2009. PMID: 18843300
-
Genes for two multicopper proteins required for Fe(III) oxide reduction in Geobacter sulfurreducens have different expression patterns both in the subsurface and on energy-harvesting electrodes.Microbiology (Reading). 2008 May;154(Pt 5):1422-1435. doi: 10.1099/mic.0.2007/014365-0. Microbiology (Reading). 2008. PMID: 18451051
-
Geobacter: the microbe electric's physiology, ecology, and practical applications.Adv Microb Physiol. 2011;59:1-100. doi: 10.1016/B978-0-12-387661-4.00004-5. Adv Microb Physiol. 2011. PMID: 22114840 Review.
-
Dissimilatory Fe(III) and Mn(IV) reduction.Adv Microb Physiol. 2004;49:219-86. doi: 10.1016/S0065-2911(04)49005-5. Adv Microb Physiol. 2004. PMID: 15518832 Review.
Cited by
-
Metagenomic applications in environmental monitoring and bioremediation.J Ind Microbiol Biotechnol. 2016 Oct;43(10):1345-54. doi: 10.1007/s10295-016-1809-8. Epub 2016 Aug 24. J Ind Microbiol Biotechnol. 2016. PMID: 27558781 Review.
-
Potential for Methanosarcina to Contribute to Uranium Reduction during Acetate-Promoted Groundwater Bioremediation.Microb Ecol. 2018 Oct;76(3):660-667. doi: 10.1007/s00248-018-1165-5. Epub 2018 Mar 2. Microb Ecol. 2018. PMID: 29500492 Free PMC article.
-
Meta-Transcriptomes From Microcosms From a Cr Impacted Soil Provides Insights Into the Metabolic Response of the Microbial Populations to Acetate Stimulation.Environ Microbiol Rep. 2025 Aug;17(4):e70148. doi: 10.1111/1758-2229.70148. Environ Microbiol Rep. 2025. PMID: 40624790 Free PMC article.
-
Metatranscriptomic Evidence for Direct Interspecies Electron Transfer between Geobacter and Methanothrix Species in Methanogenic Rice Paddy Soils.Appl Environ Microbiol. 2017 Apr 17;83(9):e00223-17. doi: 10.1128/AEM.00223-17. Print 2017 May 1. Appl Environ Microbiol. 2017. PMID: 28258137 Free PMC article.
-
Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation.ISME J. 2013 Feb;7(2):370-83. doi: 10.1038/ismej.2012.109. Epub 2012 Oct 4. ISME J. 2013. PMID: 23038171 Free PMC article.
References
-
- Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. 2009. Protists are microbes too: a perspective. ISME J. 3:4–12 - PubMed
-
- Tuomi P, Kuuppo P. 1999. Viral lysis and grazing loss of bacteria in nutrient- and carbon-manipulated brackish water enclosures. J. Plankton Res. 21:923–937
-
- Fuhrman JA, Azam F. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters—evaluation and field results. Marine Biol. 66:109–120
-
- Baath E. 1998. Growth rates of bacterial communities in soils at varying pH: a comparison of the thymidine and leucine incorporation techniques. Microb. Ecol. 36:316–327 - PubMed
-
- Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321–346 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

