rDNA-directed integration by an HIV-1 integrase--I-PpoI fusion protein
- PMID: 23275537
- PMCID: PMC3597653
- DOI: 10.1093/nar/gks1438
rDNA-directed integration by an HIV-1 integrase--I-PpoI fusion protein
Abstract
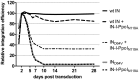
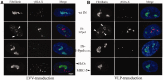
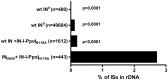
Integrating viral vectors are efficient gene transfer tools, but their integration patterns have been associated with genotoxicity and oncogenicity. The recent development of highly specific designer nucleases has enabled target DNA modification and site-specific gene insertion at desired genomic loci. However, a lack of consensus exists regarding a perfect genomic safe harbour (GSH) that would allow transgenes to be stably and reliably expressed without adversely affecting endogenous gene structure and function. Ribosomal DNA (rDNA) has many advantages as a GSH, but efficient means to target integration to this locus are currently lacking. We tested whether lentivirus vector integration can be directed to rDNA by using fusion proteins consisting of the Human Immunodeficiency Virus 1 (HIV-1) integrase (IN) and the homing endonuclease I-PpoI, which has natural cleavage sites in the rDNA. A point mutation (N119A) was introduced into I-PpoI to abolish unwanted DNA cleavage by the endonuclease. The vector-incorporated IN-I-PpoIN119A fusion protein targeted integration into rDNA significantly more than unmodified lentivirus vectors, with an efficiency of 2.7%. Our findings show that IN-fusion proteins can be used to modify the integration pattern of lentivirus vectors, and to package site-specific DNA-recognizing proteins into vectors to obtain safer transgene integration.
Figures



References
-
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. - PubMed
-
- Mussolino C, Cathomen T. TALE nucleases: tailored genome engineering made easy. Current Opin. Biotechnol. 2012;23:644–650. - PubMed
-
- Collin J, Lako M. Concise review: putting a finger on stem cell biology: zinc finger nuclease-driven targeted genetic editing in human pluripotent stem cells. Stem Cells. 2011;29:1021–1033. - PubMed

