Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast
- PMID: 23275543
- PMCID: PMC3575806
- DOI: 10.1093/nar/gks1313
Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast
Abstract
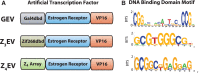
A general method for the dynamic control of single gene expression in eukaryotes, with no off-target effects, is a long-sought tool for molecular and systems biologists. We engineered two artificial transcription factors (ATFs) that contain Cys(2)His(2) zinc-finger DNA-binding domains of either the mouse transcription factor Zif268 (9 bp of specificity) or a rationally designed array of four zinc fingers (12 bp of specificity). These domains were expressed as fusions to the human estrogen receptor and VP16 activation domain. The ATFs can rapidly induce a single gene driven by a synthetic promoter in response to introduction of an otherwise inert hormone with no detectable off-target effects. In the absence of inducer, the synthetic promoter is inactive and the regulated gene product is not detected. Following addition of inducer, transcripts are induced >50-fold within 15 min. We present a quantitative characterization of these ATFs and provide constructs for making their implementation straightforward. These new tools allow for the elucidation of regulatory network elements dynamically, which we demonstrate with a major metabolic regulator, Gcn4p.
Figures





References
-
- Romanos MA, Scorer CA, Clare JJ. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. - PubMed
-
- Labbe S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Expr. Recombin. Genes Eukary. Sys. 1999;306:145–153. - PubMed
-
- Ronicke V, Graulich W, Mumberg D, Muller R, Funk M. Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Cell Cycle Control. 1997;283:313–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

