Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export
- PMID: 23275560
- PMCID: PMC3575797
- DOI: 10.1093/nar/gks1314
Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export
Abstract
We previously showed that mRNAs synthesized from three genes that naturally lack introns contain a portion of their coding sequence, known as a cytoplasmic accumulation region (CAR), which is essential for stable accumulation of the intronless mRNAs in the cytoplasm. The CAR in each mRNA is unexpectedly large, ranging in size from ∼160 to 285 nt. Here, we identified one or more copies of a 10-nt consensus sequence in each CAR. To determine whether this element (designated CAR-E) functions in cytoplasmic accumulation of intronless mRNA, we multimerized the most conserved CAR-E and inserted it upstream of β-globin cDNA, which is normally retained/degraded in the nucleus. Significantly, the tandem CAR-E, but not its antisense counterpart, rescued cytoplasmic accumulation of β-globin cDNA transcripts. Moreover, dinucleotide mutations in the CAR-E abolished this rescue. We show that the CAR-E, but not the mutant CAR-E, associates with components of the TREX mRNA export machinery, the Prp19 complex and U2AF2. Moreover, knockdown of these factors results in nuclear retention of the intronless mRNAs. Together, these data suggest that the CAR-E promotes export of intronless mRNA by sequence-dependent recruitment of the mRNA export machinery.
Figures
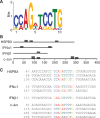
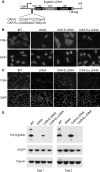
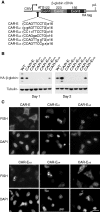
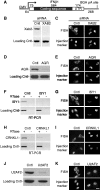
References
-
- Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogues G, Kornblihtt AR. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. - PubMed
-
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

