Network biology approach for identifying key regulatory genes by expression based study of breast cancer
- PMID: 23275709
- PMCID: PMC3530881
- DOI: 10.6026/97320630081132
Network biology approach for identifying key regulatory genes by expression based study of breast cancer
Abstract
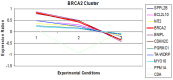
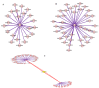
The use of high-throughput array technology is omnipresent in diverse areas specifically, early diagnosis of disease, discovery of infectious agents, search for biological markers and screening of potential drug candidates. Here, we integrated gene expression data with the network-based approach to identify novel genes that were playing central role in the network through interconnecting to a number of differentially expressed breast cancer genes. The 62 cancerous genes retrieved from the Breast Cancer Gene Database (BCGD) were mapped in the normalized data accessed from Stanford Microarray Database (SMD) to analyze their pattern. Interaction networks for each gene were constructed to understand the biology of the metastasis at systems level. The individual networks were fused together for the detection of interacting hubs, 38 novel genes were found to be deeply intermingled with the central hub node. Gene Ontology studies were made to depict the biology of the hub nodes not alone through gene ranking but by applying the Hyper geometric test with the Benjamini Hochberg False Discovery Rate (FDR) correction method at a significance level of 0.05. Analyzing p-values from the statistical test indicated that most of the novel genes were involved in the same biological function as the disordered genes like signal transducer, transcription regulator, enzyme binding, molecular transducer and receptor signaling protein activity and same pathway as MAPK signaling, Apoptosis, Wnt Signaling, ErbB signaling and Cell Cycle. Lastly, we identified 3 novel genes CHUK, INSR and CREBBP showing high connections with the 12 novel genes reported in literatures as well with the perturbed genes. As a result, these genes can be considered as significant finding in revealing the basis and pathways responsible for breast cancer.
Keywords: Breast cancer gene database (BCGD); Estrogen receptors (ER); Expression pattern; Microarray; Molecular interaction networks; Novel genes; Tamoxifen.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
