Multiple mechanisms contribute to lateral transfer of an organophosphate degradation (opd) island in Sphingobium fuliginis ATCC 27551
- PMID: 23275877
- PMCID: PMC3516476
- DOI: 10.1534/g3.112.004051
Multiple mechanisms contribute to lateral transfer of an organophosphate degradation (opd) island in Sphingobium fuliginis ATCC 27551
Abstract
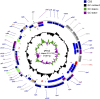
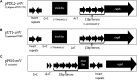
The complete sequence of pPDL2 (37,317 bp), an indigenous plasmid of Sphingobium fuliginis ATCC 27551 that encodes genes for organophosphate degradation (opd), revealed the existence of a site-specific integrase (int) gene with an attachment site attP, typically seen in integrative mobilizable elements (IME). In agreement with this sequence information, site-specific recombination was observed between pPDL2 and an artificial plasmid having a temperature-sensitive replicon and a cloned attB site at the 3' end of the seryl tRNA gene of Sphingobium japonicum. The opd gene cluster on pPDL2 was found to be part of an active catabolic transposon with mobile elements y4qE and Tn3 at its flanking ends. Besides the previously reported opd cluster, this transposon contains genes coding for protocatechuate dioxygenase and for two transport proteins from the major facilitator family that are predicted to be involved in transport and metabolism of aromatic compounds. A pPDL2 derivative, pPDL2-K, was horizontally transferred into Escherichia coli and Acinetobacter strains, suggesting that the oriT identified in pPDL2 is functional. A well-defined replicative origin (oriV), repA was identified along with a plasmid addiction module relB/relE that would support stable maintenance of pPDL2 in Sphingobium fuliginis ATCC 27551. However, if pPDL2 is laterally transferred into hosts that do not support its replication, the opd cluster appears to integrate into the host chromosome, either through transposition or through site-specific integration. The data presented in this study help to explain the existence of identical opd genes among soil bacteria.
Keywords: catabolic transposons; genomic islands; integrative conjugative elements (ICE); organophosphates; phosphotriesterase (PTE).
Figures



References
-
- Afriat L., Roodveldt C., Manco G., Tawfik D. S., 2006. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 45: 13677–13686 - PubMed
-
- Benning M. M., Kuo J. M., Raushel F. M., Holden H. M., 1994. Three-dimensional structure of phosphotriesterase: an enzyme capable of detoxifying organophosphate nerve agents. Biochemistry 33: 15001–15007 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
