A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling
- PMID: 23275879
- PMCID: PMC3516478
- DOI: 10.1534/g3.112.003780
A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling
Abstract
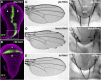
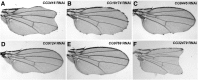
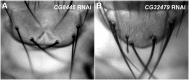
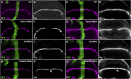
Notch signaling is highly conserved in all metazoan animals and plays critical roles in cell fate specification, cell proliferation, apoptosis, and stem cell maintenance. Although core components of the Notch signaling cascade have been identified, many gaps in the understanding of the Notch signaling pathway remain to be filled. One form of posttranslational regulation, which is controlled by the ubiquitin-proteasome system, is known to modulate Notch signaling. The ubiquitination pathway is a highly coordinated process in which the ubiquitin moiety is either conjugated to or removed from target proteins by opposing E3 ubiquitin ligases and deubiquitinases (DUBs). Several E3 ubiquitin ligases have been implicated in ubiquitin conjugation to the receptors and the ligands of the Notch signaling cascade. In contrast, little is known about a direct role of DUBs in Notch signaling in vivo. Here, we report an in vivo RNA interference screen in Drosophila melanogaster targeting all 45 DUBs that we annotated in the fly genome. We show that at least four DUBs function specifically in the formation of the fly wing margin and/or the specification of the scutellar sensory organ precursors, two processes that are strictly dependent on the balanced Notch signaling activity. Furthermore, we provide genetic evidence suggesting that these DUBs are necessary to positively modulate Notch signaling activity. Our study reveals a conserved molecular mechanism by which protein deubiquitination process contributes to the complex posttranslational regulation of Notch signaling in vivo.
Keywords: Drosophila melanogaster; Notch signaling; deubiquitinase; ubiquitination.
Figures

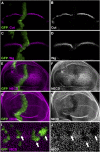
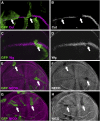
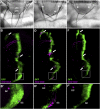
Similar articles
-
In vivo RNAi screen reveals neddylation genes as novel regulators of Hedgehog signaling.PLoS One. 2011;6(9):e24168. doi: 10.1371/journal.pone.0024168. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931660 Free PMC article.
-
Rabex-5 E3 and Rab5 GEF domains differ in their regulation of Ras, Notch, and PI3K signaling in Drosophila wing development.PLoS One. 2024 Oct 28;19(10):e0312274. doi: 10.1371/journal.pone.0312274. eCollection 2024. PLoS One. 2024. PMID: 39466792 Free PMC article.
-
A genetic mosaic screen identifies genes modulating Notch signaling in Drosophila.PLoS One. 2018 Sep 20;13(9):e0203781. doi: 10.1371/journal.pone.0203781. eCollection 2018. PLoS One. 2018. PMID: 30235233 Free PMC article.
-
The multiple facets of ubiquitination in the regulation of notch signaling pathway.Traffic. 2011 Feb;12(2):149-61. doi: 10.1111/j.1600-0854.2010.01126.x. Epub 2010 Oct 29. Traffic. 2011. PMID: 21029288 Review.
-
Regulation of Notch signaling by E3 ubiquitin ligases.FEBS J. 2022 Feb;289(4):937-954. doi: 10.1111/febs.15792. Epub 2021 Mar 16. FEBS J. 2022. PMID: 33644958 Review.
Cited by
-
Piwi reduction in the aged niche eliminates germline stem cells via Toll-GSK3 signaling.Nat Commun. 2020 Jun 19;11(1):3147. doi: 10.1038/s41467-020-16858-6. Nat Commun. 2020. PMID: 32561720 Free PMC article.
-
The Ubiquitin Conjugating Enzyme UbcD1 is Required for Notch Signaling Activation During Drosophila Wing Development.Front Genet. 2021 Oct 12;12:770853. doi: 10.3389/fgene.2021.770853. eCollection 2021. Front Genet. 2021. PMID: 34712275 Free PMC article.
-
Proteasome, but not autophagy, disruption results in severe eye and wing dysmorphia: a subunit- and regulator-dependent process in Drosophila.PLoS One. 2013 Nov 25;8(11):e80530. doi: 10.1371/journal.pone.0080530. eCollection 2013. PLoS One. 2013. PMID: 24282550 Free PMC article.
-
Cullin-4 regulates Wingless and JNK signaling-mediated cell death in the Drosophila eye.Cell Death Dis. 2016 Dec 29;7(12):e2566. doi: 10.1038/cddis.2016.338. Cell Death Dis. 2016. PMID: 28032862 Free PMC article.
-
Deubiquitinase CYLD acts as a negative regulator of dopamine neuron survival in Parkinson's disease.Sci Adv. 2022 Apr;8(13):eabh1824. doi: 10.1126/sciadv.abh1824. Epub 2022 Apr 1. Sci Adv. 2022. PMID: 35363524 Free PMC article.
References
-
- Ahmed A., Chandra S., Magarinos M., Vaessin H., 2003. Echinoid mutants exhibit neurogenic phenotypes and show synergistic interactions with the Notch signaling pathway. Development 130: 6295–6304 - PubMed
-
- Andersson E. R., Sandberg R., Lendahl U., 2011. Notch signaling: simplicity in design, versatility in function. Development 138: 3593–3612 - PubMed
-
- Artavanis-Tsakonas S., Rand M. D., Lake R. J., 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 - PubMed
-
- Blair S. S., 2007. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23: 293–319 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
