Nuclear routing networks span between nuclear pore complexes and genomic DNA to guide nucleoplasmic trafficking of biomolecules
- PMID: 23275893
- PMCID: PMC3530191
- DOI: 10.4172/2165-7491.1000112
Nuclear routing networks span between nuclear pore complexes and genomic DNA to guide nucleoplasmic trafficking of biomolecules
Abstract
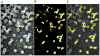
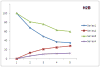
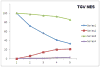
In health and disease, biomolecules, which are involved in gene expression, recombination, or reprogramming have to traffic through the nucleoplasm, between nuclear pore complexes (NPCs) and genomic DNA (gDNA). This trafficking is guided by the recently revealed nuclear routing networks (NRNs).In this study, we aimed to investigate, if the NRNs have established associations with the genomic DNA in situ and if the NRNs have capabilities to bind the DNA de novo. Moreover, we aimed to study further, if nucleoplasmic trafficking of the histones, rRNA, and transgenes' vectors, between the NPCs and gDNA, is guided by the NRNs.We used Xenopus laevis oocytes as the model system. We engineered the transgenes' DNA vectors equipped with the SV40 LTA nuclear localization signals (NLS) and/or HIV Rev nuclear export signals (NES). We purified histones, 5S rRNA, and gDNA. We rendered all these molecules superparamagnetic and fluorescent for detection with nuclear magnetic resonance (NMR), total reflection x-ray fluorescence (TXRF), energy dispersive x-ray spectroscopy (EDXS), and electron energy loss spectroscopy (EELS).The NRNs span between the NPCs and genomic DNA. They form firm bonds with the gDNA in situ. After complete digestion of the nucleic acids with the RNases and DNases, the newly added DNA - modified with the dNTP analogs, bonds firmly to the NRNs. Moreover, the NRNs guide the trafficking of the DNA transgenes' vectors - modified with the SV40 LTA NLS, following their import into the nuclei through the NPCs. The pathway is identical to that of histones. The NRNs also guide the trafficking of the DNA transgenes' vectors, modified with the HIV Rev NES, to the NPCs, followed by their export out of the nuclei. Ribosomal RNAs follow the same pathway.To summarize, the NRNs are the structures connecting the NPCs and the gDNA. They guide the trafficking of the biomolecules between the NPCs and the gDNA.
Conflict of interest statement
The authors declare no conflict of interest. The authors own the IP pertinent to this work, while protected at the WIPO and USPTO.
Figures









Similar articles
-
Routing of Biomolecules and Transgenes' Vectors in Nuclei of Oocytes.J Fertili In Vitro. 2012 Apr 30;2012(2):108-118. doi: 10.4172/2165-74. J Fertili In Vitro. 2012. PMID: 22896814 Free PMC article.
-
Nuclear Export Signal Masking Regulates HIV-1 Rev Trafficking and Viral RNA Nuclear Export.J Virol. 2017 Jan 18;91(3):e02107-16. doi: 10.1128/JVI.02107-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27852860 Free PMC article.
-
Role of nuclear pore complex in simian virus 40 nuclear targeting.J Virol. 1993 Jan;67(1):119-30. doi: 10.1128/JVI.67.1.119-130.1993. J Virol. 1993. PMID: 8380067 Free PMC article.
-
The role of nuclear envelope calcium in modifying nuclear pore complex structure.Can J Physiol Pharmacol. 2006 Mar-Apr;84(3-4):309-18. doi: 10.1139/y05-109. Can J Physiol Pharmacol. 2006. PMID: 16902578 Review.
-
Distinct nuclear import and export pathways mediated by members of the karyopherin beta family.J Cell Biochem. 1998 Aug 1;70(2):231-9. J Cell Biochem. 1998. PMID: 9671229 Review.
Cited by
-
Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication.Viruses. 2013 Nov 18;5(11):2767-95. doi: 10.3390/v5112767. Viruses. 2013. PMID: 24253283 Free PMC article. Review.
-
Eradication of Human Ovarian Cancer Cells by Transgenic Expression of Recombinant DNASE1, DNASE1L3, DNASE2, and DFFB Controlled by EGFR Promoter: Novel Strategy for Targeted Therapy of Cancer.J Genet Syndr Gene Ther. 2013 Jul 21;4(6):152. doi: 10.4172/2157-7412.1000152. J Genet Syndr Gene Ther. 2013. PMID: 24587967 Free PMC article.
References
-
- Turan S, Bode J. Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications. FASEB J. 2011;25(12):4088–107. - PubMed
-
- Hirano N, Muroi T, Takahashi H, Haruki M. Site-specific recombinases as tools for heterologous gene integration. Appl Microbiol Biotechnol. 2011;92(2):227–39. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
