Assessment of Anopheles salivary antigens as individual exposure biomarkers to species-specific malaria vector bites
- PMID: 23276246
- PMCID: PMC3547717
- DOI: 10.1186/1475-2875-11-439
Assessment of Anopheles salivary antigens as individual exposure biomarkers to species-specific malaria vector bites
Abstract
Background: Malaria transmission occurs during the blood feeding of infected anopheline mosquitoes concomitant with a saliva injection into the vertebrate host. In sub-Saharan Africa, most malaria transmission is due to Anopheles funestus s.s and to Anopheles gambiae s.l. (mainly Anopheles gambiae s.s. and Anopheles arabiensis). Several studies have demonstrated that the immune response against salivary antigens could be used to evaluate individual exposure to mosquito bites. The aim of this study was to assess the use of secreted salivary proteins as specific biomarkers of exposure to An. gambiae and/or An. funestus bites.
Methods: For this purpose, salivary gland proteins 6 (SG6) and 5'nucleotidases (5'nuc) from An. gambiae (gSG6 and g-5'nuc) and An. funestus (fSG6 and f-5'nuc) were selected and produced in recombinant form. The specificity of the IgG response against these salivary proteins was tested using an ELISA with sera from individuals living in three Senegalese villages (NDiop, n = 50; Dielmo, n = 38; and Diama, n = 46) that had been exposed to distinct densities and proportions of the Anopheles species. Individuals who had not been exposed to these tropical mosquitoes were used as controls (Marseille, n = 45).
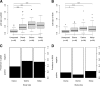
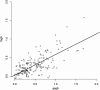
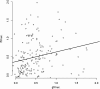
Results: The IgG responses against SG6 recombinant proteins from these two Anopheles species and against g-5'nucleotidase from An. gambiae, were significantly higher in Senegalese individuals compared with controls who were not exposed to specific Anopheles species. Conversely, an association was observed between the level of An. funestus exposure and the serological immune response levels against the f-5'nucleotidase protein.
Conclusion: This study revealed an Anopheles salivary antigenic protein that could be considered to be a promising antigenic marker to distinguish malaria vector exposure at the species level. The epidemiological interest of such species-specific antigenic markers is discussed.
Figures



References
-
- WHO. 2010. http://www.who.int.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

