Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases
- PMID: 23277537
- PMCID: PMC3549070
- DOI: 10.1073/pnas.1207104110
Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases
Abstract
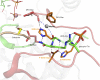
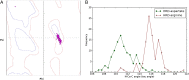
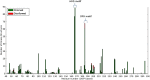
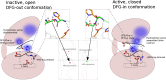
The protein kinase catalytic domain contains several conserved residues of unknown functions. Here, using a combination of computational and experimental approaches, we show that the function of some of these residues is to maintain the backbone geometry of the active site in a strained conformation. Specifically, we find that the backbone geometry of the catalytically important HRD motif deviates from ideality in high-resolution structures and the strained geometry results in favorable hydrogen bonds with conserved noncatalytic residues in the active site. In particular, a conserved aspartate in the F-helix hydrogen bonds to the strained HRD backbone in diverse eukaryotic and eukaryotic-like protein kinase crystal structures. Mutations that alter this hydrogen-bonding interaction impair catalytic activity in Aurora kinase. Although the backbone strain is present in most active conformations, several inactive conformations lack the strain because of a peptide flip in the HRD backbone. The peptide flip is correlated with loss of hydrogen bonds with the F-helix aspartate as well as with other interactions associated with kinase regulation. Within protein kinases that are regulated by activation loop phosphorylation, the strained residue is an arginine, which coordinates with the activation loop phosphate. Based on analysis of strain across the protein kinase superfamily, we propose a model in which backbone strain co-evolved with conserved residues for allosteric control of catalytic activity. Our studies provide new clues for the design of allosteric protein kinase inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101(8):2209–2242. - PubMed
-
- Krebs EG. Historical perspectives on protein phosphorylation and a classification system for protein kinases. Philos Trans R Soc Lond B Biol Sci. 1983;302(1108):3–11. - PubMed
-
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

