DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation
- PMID: 23277577
- PMCID: PMC3549119
- DOI: 10.1073/pnas.1212070110
DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation
Abstract
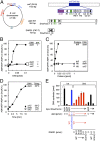
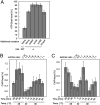
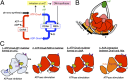
The initiation of chromosomal DNA replication is rigidly regulated to ensure that it occurs in a cell cycle-coordinated manner. To ensure this in Escherichia coli, multiple systems regulate the activity of the replication initiator ATP-DnaA. The level of ATP-DnaA increases before initiation after which it drops via DnaA-ATP hydrolysis, yielding initiation-inactive ADP-DnaA. DnaA-ATP hydrolysis is crucial to regulation of initiation and mainly occurs by a replication-coupled feedback mechanism named RIDA (regulatory inactivation of DnaA). Here, we report a second DnaA-ATP hydrolysis system that occurs at the chromosomal site datA. This locus has been annotated as a reservoir for DnaA that binds many DnaA molecules in a manner dependent upon the nucleoid-associated factor IHF (integration host factor), resulting in repression of untimely initiations; however, there is no direct evidence for the binding of many DnaA molecules at this locus. We reveal that a complex consisting of datA and IHF promotes DnaA-ATP hydrolysis in a manner dependent on specific inter-DnaA interactions. Deletion of datA or the ihf gene increased ATP-DnaA levels to the maximal attainable levels in RIDA-defective cells. Cell-cycle analysis suggested that IHF binds to datA just after replication initiation at a time when RIDA is activated. We propose a model in which cell cycle-coordinated ATP-DnaA inactivation is regulated in a concerted manner by RIDA and datA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Negative feedback for DARS2-Fis complex by ATP-DnaA supports the cell cycle-coordinated regulation for chromosome replication.Nucleic Acids Res. 2021 Dec 16;49(22):12820-12835. doi: 10.1093/nar/gkab1171. Nucleic Acids Res. 2021. PMID: 34871419 Free PMC article.
-
IHF and Fis as Escherichia coli Cell Cycle Regulators: Activation of the Replication Origin oriC and the Regulatory Cycle of the DnaA Initiator.Int J Mol Sci. 2023 Jul 18;24(14):11572. doi: 10.3390/ijms241411572. Int J Mol Sci. 2023. PMID: 37511331 Free PMC article. Review.
-
Cooperative DnaA Binding to the Negatively Supercoiled datA Locus Stimulates DnaA-ATP Hydrolysis.J Biol Chem. 2017 Jan 27;292(4):1251-1266. doi: 10.1074/jbc.M116.762815. Epub 2016 Dec 9. J Biol Chem. 2017. PMID: 27941026 Free PMC article.
-
Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation.Nucleic Acids Res. 2014 Dec 1;42(21):13134-49. doi: 10.1093/nar/gku1051. Epub 2014 Nov 6. Nucleic Acids Res. 2014. PMID: 25378325 Free PMC article.
-
Inactivation of Escherichia coli DnaA protein by DNA polymerase III and negative regulations for initiation of chromosomal replication.Biochimie. 1999 Aug-Sep;81(8-9):835-40. doi: 10.1016/s0300-9084(99)00213-8. Biochimie. 1999. PMID: 10572296 Review.
Cited by
-
Inhibition of Escherichia coli chromosome replication by rifampicin treatment or during the stringent response is overcome by de novo DnaA protein synthesis.Mol Microbiol. 2020 Dec;114(6):906-919. doi: 10.1111/mmi.14531. Epub 2020 Jun 15. Mol Microbiol. 2020. PMID: 32458540 Free PMC article.
-
Nucleotide-Induced Conformational Changes in Escherichia coli DnaA Protein Are Required for Bacterial ORC to Pre-RC Conversion at the Chromosomal Origin.Int J Mol Sci. 2015 Nov 24;16(11):27897-911. doi: 10.3390/ijms161126064. Int J Mol Sci. 2015. PMID: 26610483 Free PMC article.
-
Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli.Nucleic Acids Res. 2017 Dec 1;45(21):12354-12373. doi: 10.1093/nar/gkx914. Nucleic Acids Res. 2017. PMID: 29040689 Free PMC article.
-
The DnaA Tale.Front Microbiol. 2018 Feb 28;9:319. doi: 10.3389/fmicb.2018.00319. eCollection 2018. Front Microbiol. 2018. PMID: 29541066 Free PMC article. Review.
-
Polyphosphate induces the proteolysis of ADP-bound fraction of initiator to inhibit DNA replication initiation upon stress in Escherichia coli.Nucleic Acids Res. 2020 Jun 4;48(10):5457-5466. doi: 10.1093/nar/gkaa217. Nucleic Acids Res. 2020. PMID: 32282902 Free PMC article.
References
-
- Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26(4):355–374. - PubMed
-
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8(3):163–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

