Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing
- PMID: 23277585
- PMCID: PMC3549136
- DOI: 10.1073/pnas.1217107110
Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing
Abstract
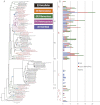
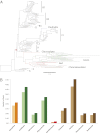
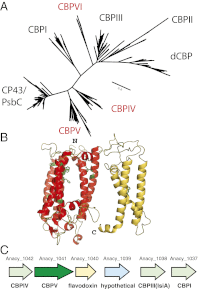
The cyanobacterial phylum encompasses oxygenic photosynthetic prokaryotes of a great breadth of morphologies and ecologies; they play key roles in global carbon and nitrogen cycles. The chloroplasts of all photosynthetic eukaryotes can trace their ancestry to cyanobacteria. Cyanobacteria also attract considerable interest as platforms for "green" biotechnology and biofuels. To explore the molecular basis of their different phenotypes and biochemical capabilities, we sequenced the genomes of 54 phylogenetically and phenotypically diverse cyanobacterial strains. Comparison of cyanobacterial genomes reveals the molecular basis for many aspects of cyanobacterial ecophysiological diversity, as well as the convergence of complex morphologies without the acquisition of novel proteins. This phylum-wide study highlights the benefits of diversity-driven genome sequencing, identifying more than 21,000 cyanobacterial proteins with no detectable similarity to known proteins, and foregrounds the diversity of light-harvesting proteins and gene clusters for secondary metabolite biosynthesis. Additionally, our results provide insight into the distribution of genes of cyanobacterial origin in eukaryotic nuclear genomes. Moreover, this study doubles both the amount and the phylogenetic diversity of cyanobacterial genome sequence data. Given the exponentially growing number of sequenced genomes, this diversity-driven study demonstrates the perspective gained by comparing disparate yet related genomes in a phylum-wide context and the insights that are gained from it.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Garcia-Pichel F, Belnap J, Neuer S, Schanz F. Estimates of global cyanobacterial biomass and its distribution. Algol Stud. 2003;109:213–227.
-
- Zehr JP, et al. Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science. 2008;322(5904):1110–1112. - PubMed
-
- Welker M, von Döhren H. Cyanobacterial peptides—nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. - PubMed
-
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

