Alphaherpesvirinae and Gammaherpesvirinae glycoprotein L and CMV UL130 originate from chemokines
- PMID: 23279912
- PMCID: PMC3598415
- DOI: 10.1186/1743-422X-10-1
Alphaherpesvirinae and Gammaherpesvirinae glycoprotein L and CMV UL130 originate from chemokines
Abstract
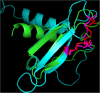
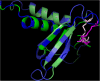
Herpesviridae is a large family of DNA viruses divided into three subfamilies: Alpha-, Beta- and Gammaherpesvirinae. The process of herpesvirus transmission is mediated by a range of proteins, one of which is glycoprotein L (gL). Based on our analysis of the solved structures of HSV2 and EBV gH/gL complexes, we propose that Alphaherpesvirinae and Gammaherpesvirinae glycoprotein L and Betaherpesvirinae UL130 originate from chemokines. Herpes simplex virus type 2 gL and human cytomegalovirus homolog (UL130) adopt a novel C chemokine-like fold, while Epstein-Barr virus gL mimics a CC chemokine structure. Hence, it is possible that gL interface with specific chemokine receptors during the transmission of Herpesviridae. We conclude that the further understanding of the function of viral chemokine-like proteins in Herpesviridae infection may lead to development of novel prophylactic and therapeutic treatment.
Figures


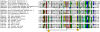
Similar articles
-
Herpes glycoprotein gL is distantly related to chemokine receptor ligands.Antiviral Res. 2007 Jul;75(1):83-6. doi: 10.1016/j.antiviral.2006.11.015. Epub 2006 Dec 26. Antiviral Res. 2007. PMID: 17215049
-
Molecular evolution of herpesviruses: genomic and protein sequence comparisons.J Virol. 1994 Mar;68(3):1886-902. doi: 10.1128/JVI.68.3.1886-1902.1994. J Virol. 1994. PMID: 8107249 Free PMC article.
-
Epstein-Barr Virus gH/gL and Kaposi's Sarcoma-Associated Herpesvirus gH/gL Bind to Different Sites on EphA2 To Trigger Fusion.J Virol. 2020 Oct 14;94(21):e01454-20. doi: 10.1128/JVI.01454-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32847853 Free PMC article.
-
Does the Zinc Finger Antiviral Protein (ZAP) Shape the Evolution of Herpesvirus Genomes?Viruses. 2021 Sep 17;13(9):1857. doi: 10.3390/v13091857. Viruses. 2021. PMID: 34578438 Free PMC article. Review.
-
Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae.Arch Virol. 1994;137(3-4):209-28. doi: 10.1007/BF01309470. Arch Virol. 1994. PMID: 7944945 Review.
Cited by
-
Chemokines encoded by herpesviruses.J Leukoc Biol. 2017 Nov;102(5):1199-1217. doi: 10.1189/jlb.4RU0417-145RR. Epub 2017 Aug 28. J Leukoc Biol. 2017. PMID: 28848041 Free PMC article. Review.
-
Rat Cytomegalovirus Virion-Associated Proteins R131 and R129 Are Necessary for Infection of Macrophages and Dendritic Cells.Pathogens. 2020 Nov 19;9(11):963. doi: 10.3390/pathogens9110963. Pathogens. 2020. PMID: 33228102 Free PMC article.
-
Mouse Cytomegalovirus Differentially Exploits Cell Surface Glycosaminoglycans in a Cell Type-Dependent and MCK-2-Independent Manner.Viruses. 2019 Dec 27;12(1):31. doi: 10.3390/v12010031. Viruses. 2019. PMID: 31892128 Free PMC article.
-
gH/gL supercomplexes at early stages of herpesvirus entry.Curr Opin Virol. 2016 Jun;18:1-8. doi: 10.1016/j.coviro.2016.01.010. Epub 2016 Feb 2. Curr Opin Virol. 2016. PMID: 26849495 Free PMC article. Review.
-
Cytomegalovirus UL128 homolog mutants that form a pentameric complex produce virus with impaired epithelial and trophoblast cell tropism and altered pathogenicity in the guinea pig.Virology. 2017 Sep;509:205-221. doi: 10.1016/j.virol.2017.06.008. Epub 2017 Jun 23. Virology. 2017. PMID: 28651121 Free PMC article.
References
-
- Whitley RJ. In: Medical Microbiology. 4. Baron S, editor. Galveston (TX); 1996. Herpesviruses.
-
- Maclean CA. HSV Entry and Spread. Meth Mol Med. 1998;10:9–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

