Conserved structural motifs in the central pair complex of eukaryotic flagella
- PMID: 23281266
- PMCID: PMC3914236
- DOI: 10.1002/cm.21094
Conserved structural motifs in the central pair complex of eukaryotic flagella
Abstract
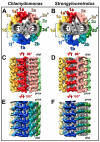
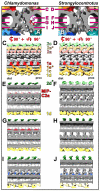
Cilia and flagella are conserved hair-like appendages of eukaryotic cells that function as sensing and motility generating organelles. Motility is driven by thousands of axonemal dyneins that require precise regulation. One essential motility regulator is the central pair complex (CPC) and many CPC defects cause paralysis of cilia/flagella. Several human diseases, such as immotile cilia syndrome, show CPC abnormalities, but little is known about the detailed three-dimensional (3D) structure and function of the CPC. The CPC is located in the center of typical [9+2] cilia/flagella and is composed of two singlet microtubules (MTs), each with a set of associated projections that extend toward the surrounding nine doublet MTs. Using cryo-electron tomography coupled with subtomogram averaging, we visualized and compared the 3D structures of the CPC in both the green alga Chlamydomonas and the sea urchin Strongylocentrotus at the highest resolution published to date. Despite the evolutionary distance between these species, their CPCs exhibit remarkable structural conservation. We identified several new projections, including those that form the elusive sheath, and show that the bridge has a more complex architecture than previously thought. Organism-specific differences include the presence of MT inner proteins in Chlamydomonas, but not Strongylocentrotus, and different overall outlines of the highly connected projection network, which forms a round-shaped cylinder in algae, but is more oval in sea urchin. These differences could be adaptations to the mechanical requirements of the rotating CPC in Chlamydomonas, compared to the Strongylocentrotus CPC which has a fixed orientation.
Copyright © 2012 Wiley Periodicals, Inc.
Figures





References
-
- Branche C, Kohl L, Toutirais G, Buisson J, Cosson J, Bastin P. Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci. 2006;119(Pt 16):3443–3455. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

