Discovering pathway cross-talks based on functional relations between pathways
- PMID: 23282018
- PMCID: PMC3521217
- DOI: 10.1186/1471-2164-13-S7-S25
Discovering pathway cross-talks based on functional relations between pathways
Abstract
Background: In biological systems, pathways coordinate or interact with one another to achieve a complex biological process. Studying how they influence each other is essential for understanding the intricacies of a biological system. However, current methods rely on statistical tests to determine pathway relations, and may lose numerous biologically significant relations.
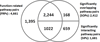
Results: This study proposes a method that identifies the pathway relations by measuring the functional relations between pathways based on the Gene Ontology (GO) annotations. This approach identified 4,661 pathway relations among 166 pathways from Pathway Interaction Database (PID). Using 143 pathway interactions from PID as testing data, the function-based approach (FBA) is able to identify 93% of pathway interactions, better than the existing methods based on the shared components and protein-protein interactions. Many well-known pathway cross-talks are only identified by FBA. In addition, the false positive rate of FBA is significantly lower than others via pathway co-expression analysis.
Conclusions: This function-based approach appears to be more sensitive and able to infer more biologically significant and explainable pathway relations.
Figures




References
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al.Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

