Phylogenetic reconstruction in the order Nymphaeales: ITS2 secondary structure analysis and in silico testing of maturase k (matK) as a potential marker for DNA bar coding
- PMID: 23282079
- PMCID: PMC3521246
- DOI: 10.1186/1471-2105-13-S17-S26
Phylogenetic reconstruction in the order Nymphaeales: ITS2 secondary structure analysis and in silico testing of maturase k (matK) as a potential marker for DNA bar coding
Abstract
Background: The Nymphaeales (waterlilly and relatives) lineage has diverged as the second branch of basal angiosperms and comprises of two families: Cabombaceae and Nymphaceae. The classification of Nymphaeales and phylogeny within the flowering plants are quite intriguing as several systems (Thorne system, Dahlgren system, Cronquist system, Takhtajan system and APG III system (Angiosperm Phylogeny Group III system) have attempted to redefine the Nymphaeales taxonomy. There have been also fossil records consisting especially of seeds, pollen, stems, leaves and flowers as early as the lower Cretaceous. Here we present an in silico study of the order Nymphaeales taking maturaseK (matK) and internal transcribed spacer (ITS2) as biomarkers for phylogeny reconstruction (using character-based methods and Bayesian approach) and identification of motifs for DNA barcoding.
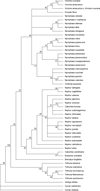
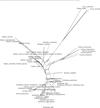
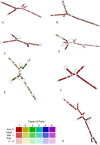
Results: The Maximum Likelihood (ML) and Bayesian approach yielded congruent fully resolved and well-supported trees using a concatenated (ITS2+ matK) supermatrix aligned dataset. The taxon sampling corroborates the monophyly of Cabombaceae. Nuphar emerges as a monophyletic clade in the family Nymphaeaceae while there are slight discrepancies in the monophyletic nature of the genera Nymphaea owing to Victoria-Euryale and Ondinea grouping in the same node of Nymphaeaceae. ITS2 secondary structures alignment corroborate the primary sequence analysis. Hydatellaceae emerged as a sister clade to Nymphaeaceae and had a basal lineage amongst the water lilly clades. Species from Cycas and Ginkgo were taken as outgroups and were rooted in the overall tree topology from various methods.
Conclusions: MatK genes are fast evolving highly variant regions of plant chloroplast DNA that can serve as potential biomarkers for DNA barcoding and also in generating primers for angiosperms with identification of unique motif regions. We have reported unique genus specific motif regions in the Order Nymphaeles from matK dataset which can be further validated for barcoding and designing of PCR primers. Our analysis using a novel approach of sequence-structure alignment and phylogenetic reconstruction using molecular morphometrics congrue with the current placement of Hydatellaceae within the early-divergent angiosperm order Nymphaeales. The results underscore the fact that more diverse genera, if not fully resolved to be monophyletic, should be represented by all major lineages.
Figures



References
-
- Borsch T, Hilu KW, Wiersema JH, Löhne C, Barthlott W, Wilde V. Phylogeny of Nymphaea (Nymphaeaceae): evidence from substitutions and microstructural changes in the chloroplast trnT-trnF region. Int J Pl Sci. 2007;168:639–671. doi: 10.1086/513476. - DOI
-
- Qiu YL, Dombrovska O, Lee J, Li L, Whitlock BA, Bernasconi-Quadroni F, Rest JS, Davis CC, Borsch T, Hilu KW, Renner SS, Soltis DE, Soltis PE, Zanis MJ, Cannone JJ, Powell M, Savolainen V, Chatrou LW, Chase MW. Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes. Int J Pl Sci. 2005;166(5):815–842. doi: 10.1086/431800. - DOI
-
- Löhne C, Borsch T, John H, Wiersema. Phylogenetic analysis of Nymphaeales using fast-evolving and noncoding chloroplast markers. Bot J Linn Soc. 2007;154(2):141–163. doi: 10.1111/j.1095-8339.2007.00659.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

