Discovery of a new class of inhibitors for the protein arginine deiminase type 4 (PAD4) by structure-based virtual screening
- PMID: 23282142
- PMCID: PMC3521205
- DOI: 10.1186/1471-2105-13-S17-S4
Discovery of a new class of inhibitors for the protein arginine deiminase type 4 (PAD4) by structure-based virtual screening
Abstract
Background: Rheumatoid arthritis (RA) is an autoimmune disease with unknown etiology. Anticitrullinated protein autoantibody has been documented as a highly specific autoantibody associated with RA. Protein arginine deiminase type 4 (PAD4) is the enzyme responsible for catalyzing the conversion of peptidylarginine into peptidylcitrulline. PAD4 is a new therapeutic target for RA treatment. In order to search for inhibitors of PAD4, structure-based virtual screening was performed using LIDAEUS (Ligand discovery at Edinburgh university). Potential inhibitors were screened experimentally by inhibition assays.
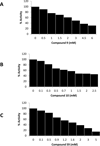
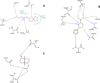
Results: Twenty two of the top-ranked water-soluble compounds were selected for inhibitory screening against PAD4. Three compounds showed significant inhibition of PAD4 and their IC50 values were investigated. The structures of the three compounds show no resemblance with previously discovered PAD4 inhibitors, nor with existing drugs for RA treatment.
Conclusion: Three compounds were discovered as potential inhibitors of PAD4 by virtual screening. The compounds are commercially available and can be used as scaffolds to design more potent inhibitors against PAD4.
Figures

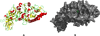


References
-
- Finesilver AG. Newer approaches to the treatment of rheumatoid arthritis. WMJ. 2003;102(7):34–37. - PubMed
-
- Aceves-Avila FJ, Medina F, Fraga A. The antiquity of rheumatoid arthritis: a reappraisal. J Rheumatol. 2001;28(4):751–757. - PubMed
-
- Rothschild BM, Coppa A, Petrone PP. "Like a virgin": absence of rheumatoid arthritis and treponematosis, good sanitation and only rare gout in Italy prior to the 15th century. Reumatismo. 2004;56(1):61–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

