PCDq: human protein complex database with quality index which summarizes different levels of evidences of protein complexes predicted from h-invitational protein-protein interactions integrative dataset
- PMID: 23282181
- PMCID: PMC3521179
- DOI: 10.1186/1752-0509-6-S2-S7
PCDq: human protein complex database with quality index which summarizes different levels of evidences of protein complexes predicted from h-invitational protein-protein interactions integrative dataset
Abstract
Background: Proteins interact with other proteins or biomolecules in complexes to perform cellular functions. Existing protein-protein interaction (PPI) databases and protein complex databases for human proteins are not organized to provide protein complex information or facilitate the discovery of novel subunits. Data integration of PPIs focused specifically on protein complexes, subunits, and their functions. Predicted candidate complexes or subunits are also important for experimental biologists.
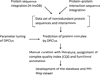
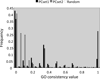
Description: Based on integrated PPI data and literature, we have developed a human protein complex database with a complex quality index (PCDq), which includes both known and predicted complexes and subunits. We integrated six PPI data (BIND, DIP, MINT, HPRD, IntAct, and GNP_Y2H), and predicted human protein complexes by finding densely connected regions in the PPI networks. They were curated with the literature so that missing proteins were complemented and some complexes were merged, resulting in 1,264 complexes comprising 9,268 proteins with 32,198 PPIs. The evidence level of each subunit was assigned as a categorical variable. This indicated whether it was a known subunit, and a specific function was inferable from sequence or network analysis. To summarize the categories of all the subunits in a complex, we devised a complex quality index (CQI) and assigned it to each complex. We examined the proportion of consistency of Gene Ontology (GO) terms among protein subunits of a complex. Next, we compared the expression profiles of the corresponding genes and found that many proteins in larger complexes tend to be expressed cooperatively at the transcript level. The proportion of duplicated genes in a complex was evaluated. Finally, we identified 78 hypothetical proteins that were annotated as subunits of 82 complexes, which included known complexes. Of these hypothetical proteins, after our prediction had been made, four were reported to be actual subunits of the assigned protein complexes.
Conclusions: We constructed a new protein complex database PCDq including both predicted and curated human protein complexes. CQI is a useful source of experimentally confirmed information about protein complexes and subunits. The predicted protein complexes can provide functional clues about hypothetical proteins. PCDq is freely available at http://h-invitational.jp/hinv/pcdq/.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

