Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species
- PMID: 23283959
- PMCID: PMC3592153
- DOI: 10.1128/JVI.02719-12
Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species
Abstract
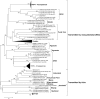
Evolutionary insights into the phleboviruses are limited because of an imprecise classification scheme based on partial nucleotide sequences and scattered antigenic relationships. In this report, the serologic and phylogenetic relationships of the Uukuniemi group viruses and their relationships with other recently characterized tick-borne phleboviruses are described using full-length genome sequences. We propose that the viruses currently included in the Uukuniemi virus group be assigned to five different species as follows: Uukuniemi virus, EgAn 1825-61 virus, Fin V707 virus, Chizé virus, and Zaliv Terpenia virus would be classified into the Uukuniemi species; Murre virus, RML-105-105355 virus, and Sunday Canyon virus would be classified into a Murre virus species; and Grand Arbaud virus, Precarious Point virus, and Manawa virus would each be given individual species status. Although limited sequence similarity was detected between current members of the Uukuniemi group and Severe fever with thrombocytopenia syndrome virus (SFTSV) and Heartland virus, a clear serological reaction was observed between some of them, indicating that SFTSV and Heartland virus should be considered part of the Uukuniemi virus group. Moreover, based on the genomic diversity of the phleboviruses and given the low correlation observed between complement fixation titers and genetic distance, we propose a system for classification of the Bunyaviridae based on genetic as well as serological data. Finally, the recent descriptions of SFTSV and Heartland virus also indicate that the public health importance of the Uukuniemi group viruses must be reevaluated.
Figures


References
-
- Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn C, Tesh R. 2005. Family Bunyaviridae. Elsevier Academic Press, London, United Kingdom
-
- Henderson WW, Monroe MC, St Jeor SC, Thayer WP, Rowe JE, Peters CJ, Nichol ST. 1995. Naturally occurring Sin Nombre virus genetic reassortants. Virology 214:602–610 - PubMed
-
- Li D, Schmaljohn AL, Anderson K, Schmaljohn CS. 1995. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology 206:973–983 - PubMed
-
- Pringle CR, Lees JF, Clark W, Elliott RM. 1984. Genome subunit reassortment among Bunyaviruses analysed by dot hybridization using molecularly cloned complementary DNA probes. Virology 135:244–256 - PubMed
-
- Rodriguez LL, Owens JH, Peters CJ, Nichol ST. 1998. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology 242:99–106 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

