Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system
- PMID: 23283983
- PMCID: PMC3583667
- DOI: 10.1091/mbc.E12-09-0659
Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system
Abstract
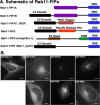
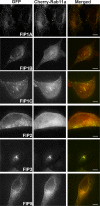
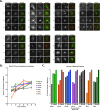
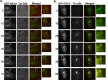
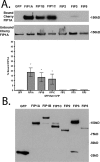
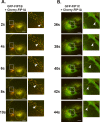
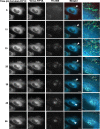
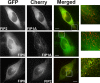
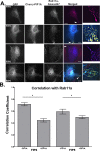
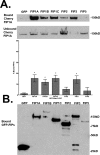
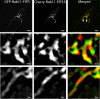
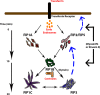
The Rab11-family interacting proteins (Rab11-FIPs) facilitate Rab11-dependent vesicle recycling. We hypothesized that Rab11-FIPs define discrete subdomains and carry out temporally distinct roles within the recycling system. We used live-cell deconvolution microscopy of HeLa cells expressing chimeric fluorescent Rab11-FIPs to examine Rab11-FIP localization, transferrin passage through Rab11-FIP-containing compartments, and overlap among Rab11-FIPs within the recycling system. FIP1A, FIP2, and FIP5 occupy widely distributed mobile tubules and vesicles, whereas FIP1B, FIP1C, and FIP3 localize to perinuclear tubules. Internalized transferrin entered Rab11-FIP-containing compartments within 5 min, reaching maximum colocalization with FIP1B and FIP2 early in the time course, whereas localization with FIP1A, FIP1C, FIP3, and FIP5 was delayed until 10 min or later. Whereas direct interactions with FIP1A were only observed for FIP1B and FIP1C, FIP1A also associated with membranes containing FIP3. Live-cell dual-expression studies of Rab11-FIPs revealed the tubular dynamics of Rab11-FIP-containing compartments and demonstrated a series of selective associations among Rab11-FIPs in real time. These findings suggest that Rab11-FIP1 proteins participate in spatially and temporally distinct steps of the recycling process along a complex and dynamic tubular network in which Rab11-FIPs occupy discrete domains.
Figures

References
-
- Anderson RG, Brown MS, Goldstein JL. Role of the coated endocytic vesicle in the uptake of receptor-bound low density lipoprotein in human fibroblasts. Cell. 1977;10:351–364. - PubMed
-
- Basu SK, Goldstein JL, Anderson RG, Brown MS. Monensin interrupts the recycling of low density lipoprotein receptors in human fibroblasts. Cell. 1981;24:493–502. - PubMed
-
- Bhartur SG, Calhoun BC, Woodrum J, Kurkjian J, Iyer S, Lai F, Goldenring JR. Genomic structure of murine Rab11 family members. Biochem Biophys Res Commun. 2000;269:611–617. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

