Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture
- PMID: 23284287
- PMCID: PMC3527209
- DOI: 10.1371/journal.pgen.1003080
Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture
Abstract
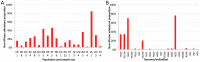
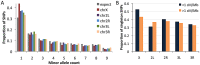
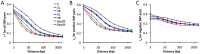
Drosophila melanogaster has played a pivotal role in the development of modern population genetics. However, many basic questions regarding the demographic and adaptive history of this species remain unresolved. We report the genome sequencing of 139 wild-derived strains of D. melanogaster, representing 22 population samples from the sub-Saharan ancestral range of this species, along with one European population. Most genomes were sequenced above 25X depth from haploid embryos. Results indicated a pervasive influence of non-African admixture in many African populations, motivating the development and application of a novel admixture detection method. Admixture proportions varied among populations, with greater admixture in urban locations. Admixture levels also varied across the genome, with localized peaks and valleys suggestive of a non-neutral introgression process. Genomes from the same location differed starkly in ancestry, suggesting that isolation mechanisms may exist within African populations. After removing putatively admixed genomic segments, the greatest genetic diversity was observed in southern Africa (e.g. Zambia), while diversity in other populations was largely consistent with a geographic expansion from this potentially ancestral region. The European population showed different levels of diversity reduction on each chromosome arm, and some African populations displayed chromosome arm-specific diversity reductions. Inversions in the European sample were associated with strong elevations in diversity across chromosome arms. Genomic scans were conducted to identify loci that may represent targets of positive selection within an African population, between African populations, and between European and African populations. A disproportionate number of candidate selective sweep regions were located near genes with varied roles in gene regulation. Outliers for Europe-Africa F(ST) were found to be enriched in genomic regions of locally elevated cosmopolitan admixture, possibly reflecting a role for some of these loci in driving the introgression of non-African alleles into African populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. (2000) The genome sequence of Drosophila melanogaster . Science 287: 2185–2195. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

