Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa
- PMID: 23284289
- PMCID: PMC3527224
- DOI: 10.1371/journal.pgen.1003093
Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa
Abstract
Genome duplication, which results in polyploidy, is disruptive to fundamental biological processes. Genome duplications occur spontaneously in a range of taxa and problems such as sterility, aneuploidy, and gene expression aberrations are common in newly formed polyploids. In mammals, genome duplication is associated with cancer and spontaneous abortion of embryos. Nevertheless, stable polyploid species occur in both plants and animals. Understanding how natural selection enabled these species to overcome early challenges can provide important insights into the mechanisms by which core cellular functions can adapt to perturbations of the genomic environment. Arabidopsis arenosa includes stable tetraploid populations and is related to well-characterized diploids A. lyrata and A. thaliana. It thus provides a rare opportunity to leverage genomic tools to investigate the genetic basis of polyploid stabilization. We sequenced the genomes of twelve A. arenosa individuals and found signatures suggestive of recent and ongoing selective sweeps throughout the genome. Many of these are at genes implicated in genome maintenance functions, including chromosome cohesion and segregation, DNA repair, homologous recombination, transcriptional regulation, and chromatin structure. Numerous encoded proteins are predicted to interact with one another. For a critical meiosis gene, ASYNAPSIS1, we identified a non-synonymous mutation that is highly differentiated by cytotype, but present as a rare variant in diploid A. arenosa, indicating selection may have acted on standing variation already present in the diploid. Several genes we identified that are implicated in sister chromatid cohesion and segregation are homologous to genes identified in a yeast mutant screen as necessary for survival of polyploid cells, and also implicated in genome instability in human diseases including cancer. This points to commonalities across kingdoms and supports the hypothesis that selection has acted on genes controlling genome integrity in A. arenosa as an adaptive response to genome doubling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
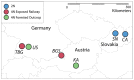
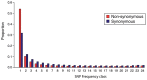


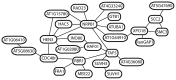
Comment in
-
Evolution: he who grabs too much loses all.Curr Biol. 2013 Nov 4;23(21):R961-3. doi: 10.1016/j.cub.2013.09.023. Curr Biol. 2013. PMID: 24200324
References
-
- Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846. - PubMed
-
- Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19: 141–147. - PubMed
-
- Otto SP (2007) The evolutionary consequences of polyploidy. Cell 131: 452–462. - PubMed
-
- Parisod C, Holderegger R, Brochmann C (2010) Evolutionary consequences of autopolyploidy. New Phytol 186: 5–17. - PubMed
-
- Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Ann Rev Ecol Systemat 33: 589–639.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

