RecX facilitates homologous recombination by modulating RecA activities
- PMID: 23284295
- PMCID: PMC3527212
- DOI: 10.1371/journal.pgen.1003126
RecX facilitates homologous recombination by modulating RecA activities
Abstract
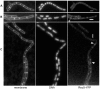
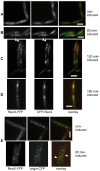
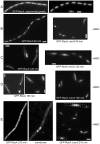
The Bacillus subtilis recH342 strain, which decreases interspecies recombination without significantly affecting the frequency of transformation with homogamic DNA, carried a point mutation in the putative recX (yfhG) gene, and the mutation was renamed as recX342. We show that RecX (264 residues long), which shares partial identity with the Proteobacterial RecX (<180 residues), is a genuine recombination protein, and its primary function is to modulate the SOS response and to facilitate RecA-mediated recombinational repair and genetic recombination. RecX-YFP formed discrete foci on the nucleoid, which were coincident in time with RecF, in response to DNA damage, and on the poles and/or the nucleoid upon stochastic induction of programmed natural competence. When DNA was damaged, the RecX foci co-localized with RecA threads that persisted for a longer time in the recX context. The absence of RecX severely impaired natural transformation both with plasmid and chromosomal DNA. We show that RecX suppresses the negative effect exerted by RecA during plasmid transformation, prevents RecA mis-sensing of single-stranded DNA tracts, and modulates DNA strand exchange. RecX, by modulating the "length or packing" of a RecA filament, facilitates the initiation of recombination and increases recombination across species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Bacillus subtilis RecA with DprA-SsbA antagonizes RecX function during natural transformation.Nucleic Acids Res. 2017 Sep 6;45(15):8873-8885. doi: 10.1093/nar/gkx583. Nucleic Acids Res. 2017. PMID: 28911099 Free PMC article.
-
Bacillus subtilis RecA and its accessory factors, RecF, RecO, RecR and RecX, are required for spore resistance to DNA double-strand break.Nucleic Acids Res. 2014 Feb;42(4):2295-307. doi: 10.1093/nar/gkt1194. Epub 2013 Nov 26. Nucleic Acids Res. 2014. PMID: 24285298 Free PMC article.
-
RecA Is Required for the Assembly of RecN into DNA Repair Complexes on the Nucleoid.J Bacteriol. 2021 Sep 23;203(20):e0024021. doi: 10.1128/JB.00240-21. Epub 2021 Aug 2. J Bacteriol. 2021. PMID: 34339298 Free PMC article.
-
[Enzymatic control of homologous recombination in Escherichia coli cells and hyper-recombination].Mol Biol (Mosk). 2013 Mar-Apr;47(2):205-17. doi: 10.7868/s0026898413020031. Mol Biol (Mosk). 2013. PMID: 23808153 Review. Russian.
-
The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery.Crit Rev Biochem Mol Biol. 2012 Nov-Dec;47(6):531-55. doi: 10.3109/10409238.2012.729562. Epub 2012 Oct 9. Crit Rev Biochem Mol Biol. 2012. PMID: 23046409 Free PMC article. Review.
Cited by
-
The RecD2 helicase balances RecA activities.Nucleic Acids Res. 2022 Apr 8;50(6):3432-3444. doi: 10.1093/nar/gkac131. Nucleic Acids Res. 2022. PMID: 35234892 Free PMC article.
-
RECX Interacts with Mitochondrial RECA to Maintain Mitochondrial Genome Stability.Plant Physiol. 2018 May;177(1):300-310. doi: 10.1104/pp.18.00218. Epub 2018 Mar 26. Plant Physiol. 2018. PMID: 29581177 Free PMC article.
-
PcrA Dissociates RecA Filaments and the SsbA and RecO Mediators Counterbalance Such Activity.Front Mol Biosci. 2022 Feb 9;9:836211. doi: 10.3389/fmolb.2022.836211. eCollection 2022. Front Mol Biosci. 2022. PMID: 35223992 Free PMC article.
-
ATPase Activity of Bacillus subtilis RecA Affects the Dynamic Formation of RecA Filaments at DNA Double Strand Breaks.mSphere. 2022 Dec 21;7(6):e0041222. doi: 10.1128/msphere.00412-22. Epub 2022 Nov 2. mSphere. 2022. PMID: 36321831 Free PMC article.
-
Single molecule tracking reveals functions for RarA at replication forks but also independently from replication during DNA repair in Bacillus subtilis.Sci Rep. 2019 Feb 13;9(1):1997. doi: 10.1038/s41598-018-38289-6. Sci Rep. 2019. PMID: 30760776 Free PMC article.
References
-
- Taddei F, Matic I, Godelle B, Radman M (1997) To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol 5: 427–428. - PubMed
-
- Matic I, Rayssiguier C, Radman M (1995) Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80: 507–515. - PubMed
-
- Delmas S, Matic I (2005) Cellular response to horizontally transferred DNA in Escherichia coli is tuned by DNA repair systems. DNA repair 4: 221–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

