Network Inference and Biological Dynamics
- PMID: 23284600
- PMCID: PMC3533376
- DOI: 10.1214/11-AOAS532
Network Inference and Biological Dynamics
Abstract
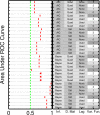
Network inference approaches are now widely used in biological applications to probe regulatory relationships between molecular components such as genes or proteins. Many methods have been proposed for this setting, but the connections and differences between their statistical formulations have received less attention. In this paper, we show how a broad class of statistical network inference methods, including a number of existing approaches, can be described in terms of variable selection for the linear model. This reveals some subtle but important differences between the methods, including the treatment of time intervals in discretely observed data. In developing a general formulation, we also explore the relationship between single-cell stochastic dynamics and network inference on averages over cells. This clarifies the link between biochemical networks as they operate at the cellular level and network inference as carried out on data that are averages over populations of cells. We present empirical results, comparing thirty-two network inference methods that are instances of the general formulation we describe, using two published dynamical models. Our investigation sheds light on the applicability and limitations of network inference and provides guidance for practitioners and suggestions for experimental design.
Figures



References
-
- Äijö T, Lähdesmäki H. Learning gene regulatory networks from gene expression measurements using non-parametric molecular kinetics. Bioinformatics. 2009;25(22):2937–44. - PubMed
-
- Altay G, Emmert- Streib F. Revealing differences in gene network inference algorithms on the network level by ensemble methods. Bioinformatics. 2010;26(14):1738–1744. - PubMed
-
- Babu MM, Luscombe NM, Aravind L, et al. Structure and evolution of transcriptional regulatory networks. Current Opinion in Structural Biology. 2004;14(3):283–91. - PubMed
-
- Bansal M, di Bernardo D. Inference of gene networks from temporal gene expression profiles. IET Systems Biology. 2007;5:306–12. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
