Using family-based imputation in genome-wide association studies with large complex pedigrees: the Framingham Heart Study
- PMID: 23284720
- PMCID: PMC3524237
- DOI: 10.1371/journal.pone.0051589
Using family-based imputation in genome-wide association studies with large complex pedigrees: the Framingham Heart Study
Abstract
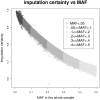
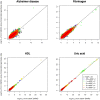
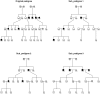
Imputation has been widely used in genome-wide association studies (GWAS) to infer genotypes of un-genotyped variants based on the linkage disequilibrium in external reference panels such as the HapMap and 1000 Genomes. However, imputation has only rarely been performed based on family relationships to infer genotypes of un-genotyped individuals. Using 8998 Framingham Heart Study (FHS) participants genotyped with Affymetrix 550K SNPs, we imputed genotypes of same set of SNPs for additional 3121 participants, most of whom were never genotyped due to lack of DNA sample. Prior to imputation, 122 pedigrees were too large to be handled by the imputation software Merlin. Therefore, we developed a novel pedigree splitting algorithm that can maximize the number of genotyped relatives for imputing each un-genotyped individual, while keeping new sub-pedigrees under a pre-specified size. In GWAS of four phenotypes available in FHS (Alzheimer disease, circulating levels of fibrinogen, high-density lipoprotein cholesterol, and uric acid), we compared results using genotyped individuals only with results using both genotyped and imputed individuals. We studied the impact of applying different imputation quality filtering thresholds on the association results and did not found a universal threshold that always resulted in a more significant p-value for previously identified loci. However most of these loci had a lower p-value when we only included imputed genotypes with with ≥60% SNP- and ≥50% person-specific imputation certainty. In summary, we developed a novel algorithm for splitting large pedigrees for imputation and found a plausible imputation quality filtering threshold based on FHS. Further examination may be required to generalize this threshold to other studies.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS017950/NS/NINDS NIH HHS/United States
- R01HL093029/HL/NHLBI NIH HHS/United States
- N01 HC025195/HC/NHLBI NIH HHS/United States
- R01 HL093029/HL/NHLBI NIH HHS/United States
- R01HL093328/HL/NHLBI NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01NS017950/NS/NINDS NIH HHS/United States
- R01 HL093328/HL/NHLBI NIH HHS/United States
- N02-HL-6-4278/HL/NHLBI NIH HHS/United States
- N01-HC-25195/HC/NHLBI NIH HHS/United States
- R01 NS017950-28/NS/NINDS NIH HHS/United States
- R01-HL093328-01/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

