Heterologous SUMO-2/3-ubiquitin chains optimize IκBα degradation and NF-κB activity
- PMID: 23284737
- PMCID: PMC3527444
- DOI: 10.1371/journal.pone.0051672
Heterologous SUMO-2/3-ubiquitin chains optimize IκBα degradation and NF-κB activity
Abstract
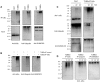
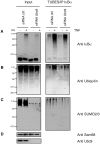
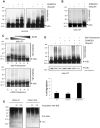
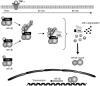
The NF-κB pathway is regulated by SUMOylation at least at three levels: the inhibitory molecule IκBα, the IKK subunit γ/NEMO and the p52 precursor p100. Here we investigate the role of SUMO-2/3 in the degradation of IκBα and activation of NF-κB mediated by TNFα. We found that under conditions of deficient SUMOylation, an important delay in both TNFα-mediated proteolysis of IκBα and NF-κB dependent transcription occurs. In vitro and ex vivo approaches, including the use of ubiquitin-traps (TUBEs), revealed the formation of chains on IκBα containing SUMO-2/3 and ubiquitin after TNFα stimulation. The integration of SUMO-2/3 appears to promote the formation of ubiquitin chains on IκBα after activation of the TNFα signalling pathway. Furthermore, heterologous chains of SUMO-2/3 and ubiquitin promote a more efficient degradation of IκBα by the 26S proteasome in vitro compared to chains of either SUMO-2/3 or ubiquitin alone. Consistently, Ubc9 silencing reduced the capture of IκBα modified with SUMO-ubiquitin hybrid chains that display a defective proteasome-mediated degradation. Thus, hybrid SUMO-2/3-ubiquitin chains increase the susceptibility of modified IκBα to the action of 26S proteasome, contributing to the optimal control of NF-κB activity after TNFα-stimulation.
Conflict of interest statement
Figures




References
-
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12. - PubMed
-
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479. - PubMed
-
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, et al. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136: 1098–1109. - PubMed
-
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, et al. (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 11: 123–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

