Identification and expression of nine oak aquaporin genes in the primary root axis of two oak species, Quercus petraea and Quercus robur
- PMID: 23284785
- PMCID: PMC3524086
- DOI: 10.1371/journal.pone.0051838
Identification and expression of nine oak aquaporin genes in the primary root axis of two oak species, Quercus petraea and Quercus robur
Abstract
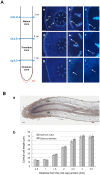
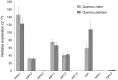
Aquaporins (AQPs) belong to the Major Intrinsic Protein family that conducts water and other small solutes across biological membranes. This study aimed to identify and characterize AQP genes in the primary root axis of two oak species, Quercus petraea and Quercus robur. Nine putative AQP genes were cloned, and their expression was profiled in different developmental root zones by real-time PCR. A detailed examination of the predicted amino acid sequences and subsequent phylogenetic analysis showed that the isolated AQPs could be divided into two subfamilies, which included six plasma membrane intrinsic proteins (PIPs) and three tonoplast intrinsic proteins (TIPs). We characterized the anatomical features of the roots and defined three developmental root zones: the immature, transition and mature zones. Expression analysis of the AQPs was performed according to these root developmental stages. Our results showed that the expression of PIP2;3 and TIP1 was significantly higher in Quercus petraea compared with Quercus robur in the three root zones. However, PIP2;1 and TIP2;1 were found to be differentially expressed in the mature zone of the two oak species. Of the nine AQP genes identified and analyzed, we highlighted four genes that might facilitate a deeper understanding of how these two closely related tree species adapted to different environments.
Conflict of interest statement
Figures



Similar articles
-
Short-term response to waterlogging in Quercus petraea and Quercus robur: A study of the root hydraulic responses and the transcriptional pattern of aquaporins.Plant Physiol Biochem. 2015 Dec;97:323-30. doi: 10.1016/j.plaphy.2015.10.016. Epub 2015 Oct 19. Plant Physiol Biochem. 2015. PMID: 26519820
-
Localization and quantification of plasma membrane aquaporin expression in maize primary root: a clue to understanding their role as cellular plumbers.Plant Mol Biol. 2006 Sep;62(1-2):305-23. doi: 10.1007/s11103-006-9022-1. Epub 2006 Jul 15. Plant Mol Biol. 2006. PMID: 16845476
-
Identification of adaptation-specific differences in mRNA expression of sessile and pedunculate oak based on osmotic-stress-induced genes.Tree Physiol. 2005 Oct;25(10):1317-29. doi: 10.1093/treephys/25.10.1317. Tree Physiol. 2005. PMID: 16076780
-
Implication of the suberin pathway in adaptation to waterlogging and hypertrophied lenticels formation in pedunculate oak (Quercus robur L.).Tree Physiol. 2016 Nov;36(11):1330-1342. doi: 10.1093/treephys/tpw056. Epub 2016 Jun 29. Tree Physiol. 2016. PMID: 27358207
-
Genome-wide comparative analysis of tonoplast intrinsic protein (TIP) genes in plants.Funct Integr Genomics. 2014 Dec;14(4):617-29. doi: 10.1007/s10142-014-0389-9. Epub 2014 Aug 6. Funct Integr Genomics. 2014. PMID: 25095751 Review.
Cited by
-
Physiological and transcriptomic analyses reveal the regulatory mechanisms for the adaptation of Quercus robur to shade conditions.BMC Plant Biol. 2025 Jul 2;25(1):821. doi: 10.1186/s12870-025-06843-w. BMC Plant Biol. 2025. PMID: 40604453 Free PMC article.
References
-
- Steudle E (2000) Water uptake by plant roots: an integration of views. Plant Soil 226: 45–56.
-
- Enstone DE, Peterson CA (2005) Suberin lamella development in maize seedling roots grown in aerated and stagnant conditions. Plant Cell Environ 28: 444–455.
-
- Alexandersson E, Danielson JAH, Rade J, Moparthi VK, Fontes M, et al. (2010) Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J 61: 650–660. - PubMed
-
- Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Ann Review of Plant Biol 59: 595–624. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

