Genotyping of fanconi anemia patients by whole exome sequencing: advantages and challenges
- PMID: 23285130
- PMCID: PMC3527584
- DOI: 10.1371/journal.pone.0052648
Genotyping of fanconi anemia patients by whole exome sequencing: advantages and challenges
Abstract
Fanconi anemia (FA) is a rare genomic instability syndrome. Disease-causing are biallelic mutations in any one of at least 15 genes encoding members of the FA/BRCA pathway of DNA-interstrand crosslink repair. Patients are diagnosed based upon phenotypical manifestations and the diagnosis of FA is confirmed by the hypersensitivity of cells to DNA interstrand crosslinking agents. Customary molecular diagnostics has become increasingly cumbersome, time-consuming and expensive the more FA genes have been identified. We performed Whole Exome Sequencing (WES) in four FA patients in order to investigate the potential of this method for FA genotyping. In search of an optimal WES methodology we explored different enrichment and sequencing techniques. In each case we were able to identify the pathogenic mutations so that WES provided both, complementation group assignment and mutation detection in a single approach. The mutations included homozygous and heterozygous single base pair substitutions and a two-base-pair duplication in FANCJ, -D1, or -D2. Different WES strategies had no critical influence on the individual outcome. However, database errors and in particular pseudogenes impose obstacles that may prevent correct data perception and interpretation, and thus cause pitfalls. With these difficulties in mind, our results show that WES is a valuable tool for the molecular diagnosis of FA and a sufficiently safe technique, capable of engaging increasingly in competition with classical genetic approaches.
Conflict of interest statement
Figures
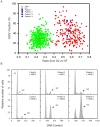

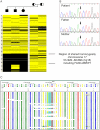
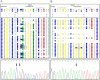

References
-
- Vuorela M, Pylkas K, Hartikainen JM, Sundfeldt K, Lindblom A, et al. (2011) Further evidence for the contribution of the RAD51C gene in hereditary breast and ovarian cancer susceptibility. Breast Cancer Res Treat 130: 1003–1010. - PubMed
-
- Alter BP, Kupfer G (1993) Fanconi Anemia. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews. Seattle (WA). - PubMed
-
- Seif AE (2011) Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis. Cancer Genet 204: 227–244. - PubMed
-
- Rosenberg PS, Greene MH, Alter BP (2003) Cancer incidence in persons with Fanconi anemia. Blood 101: 822–826. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

