Diverged alleles of the Anopheles gambiae leucine-rich repeat gene APL1A display distinct protective profiles against Plasmodium falciparum
- PMID: 23285147
- PMCID: PMC3532451
- DOI: 10.1371/journal.pone.0052684
Diverged alleles of the Anopheles gambiae leucine-rich repeat gene APL1A display distinct protective profiles against Plasmodium falciparum
Abstract
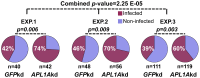
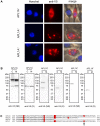
Functional studies have demonstrated a role for the Anopheles gambiae APL1A gene in resistance against the human malaria parasite, Plasmodium falciparum. Here, we exhaustively characterize the structure of the APL1 locus and show that three structurally different APL1A alleles segregate in the Ngousso colony. Genetic association combined with RNAi-mediated gene silencing revealed that APL1A alleles display distinct protective profiles against P. falciparum. One APL1A allele is sufficient to explain the protective phenotype of APL1A observed in silencing experiments. Epitope-tagged APL1A isoforms expressed in an in vitro hemocyte-like cell system showed that under assay conditions, the most protective APL1A isoform (APL1A(2)) localizes within large cytoplasmic vesicles, is not constitutively secreted, and forms only one protein complex, while a less protective isoform (APL1A(1)) is constitutively secreted in at least two protein complexes. The tested alleles are identical to natural variants in the wild A. gambiae population, suggesting that APL1A genetic variation could be a factor underlying natural heterogeneity of vector susceptibility to P. falciparum.
Conflict of interest statement
Figures



References
-
- TDR/WHO (2012) Malaria.
-
- Niare O, Markianos K, Volz J, Oduol F, Toure A, et al. (2002) Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science 298: 213–216. - PubMed
-
- Riehle MM, Markianos K, Niare O, Xu J, Li J, et al. (2006) Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 312: 577–579. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

