Rapid and serial quantification of adhesion forces of yeast and Mammalian cells
- PMID: 23285166
- PMCID: PMC3527581
- DOI: 10.1371/journal.pone.0052712
Rapid and serial quantification of adhesion forces of yeast and Mammalian cells
Abstract
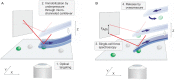
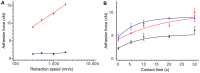
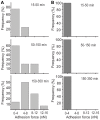
Cell adhesion to surfaces represents the basis for niche colonization and survival. Here we establish serial quantification of adhesion forces of different cell types using a single probe. The pace of single-cell force-spectroscopy was accelerated to up to 200 yeast and 20 mammalian cells per probe when replacing the conventional cell trapping cantilever chemistry of atomic force microscopy by underpressure immobilization with fluidic force microscopy (FluidFM). In consequence, statistically relevant data could be recorded in a rapid manner, the spectrum of examinable cells was enlarged, and the cell physiology preserved until approached for force spectroscopy. Adhesion forces of Candida albicans increased from below 4 up to 16 nN at 37°C on hydrophobic surfaces, whereas a Δhgc1-mutant showed forces consistently below 4 nN. Monitoring adhesion of mammalian cells revealed mean adhesion forces of 600 nN of HeLa cells on fibronectin and were one order of magnitude higher than those observed for HEK cells.
Conflict of interest statement
Figures



References
-
- Baror Y (1990) The effect of adhesion on survival and growth of microorganisms. Experientia 46: 823–826.
-
- An YH, Friedman RJ (1997) Laboratory methods for studies of bacterial adhesion. Journal of Microbiological Methods 30: 141–152.
-
- Ashkin A, Dziedzic JM (1987) Optical trapping and manipulation of viruses and bacteria. Science 235: 1517–1520. - PubMed
-
- Walter N, Selhuber C, Kessler H, Spatz JP (2006) Cellular unbinding forces of initial adhesion processes on nanopatterned surfaces probed with magnetic tweezers. Nano Letters 6: 398–402. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

