The MTA family proteins as novel histone H3 binding proteins
- PMID: 23286669
- PMCID: PMC3562248
- DOI: 10.1186/2045-3701-3-1
The MTA family proteins as novel histone H3 binding proteins
Erratum in
-
Correction: the MTA family proteins as novel histone H3 binding proteins.Cell Biosci. 2013 Feb 15;3(1):12. doi: 10.1186/2045-3701-3-12. Cell Biosci. 2013. PMID: 23414553 Free PMC article. No abstract available.
Abstract
Background: The nucleosome remodeling and histone deacetylase complex (Mi2/NRD/NuRD/NURD) has a broad role in regulation of transcription, DNA repair and cell cycle. Previous studies have revealed a specific interaction between NURD and histone H3N-terminal tail in vitro that is not observed for another HDAC1/2-containing complex, Sin3A. However, the subunit(s) responsible for specific binding of H3 by NURD has not been defined.
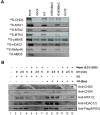
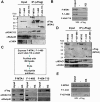
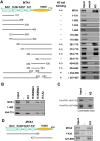
Results: In this study, we show among several class I HDAC-containing corepressor complexes only NURD exhibits a substantial H3 tail-binding activity in vitro. We present the evidence that the MTA family proteins within the NURD complex interact directly with H3 tail. Extensive in vitro binding assays mapped the H3 tail-binding domain to the C-terminal region of MTA1 and MTA2. Significantly, although the MTA1 and MTA2 mutant proteins with deletion of the C-terminal H3 tail binding domain were assembled into the endogenous NURD complex when expressed in mammalian cells, the resulting NURD complexes were deficient in binding H3 tail in vitro, indicating that the MTA family proteins are required for the observed specific binding of H3 tail peptide by NURD in vitro. However, chromatin fractionation experiments show that the NURD complexes with impaired MTA1/2-H3 tail binding activity remained to be associated with chromatin in cells.
Conclusions: Together our study reveals a novel histone H3-binding activity for the MTA family proteins and provides evidence that the MTA family proteins mediate the in vitro specific binding of H3 tail peptide by NURD complex. However, multiple mechanisms are likely to contribute to the chromatin association of NURD complex in cells. Our finding also raises the possibility that the MTA family proteins may exert their diverse biological functions at least in part through their direct interaction with H3 tail.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

