The role of chromatin modifiers in normal and malignant hematopoiesis
- PMID: 23287864
- PMCID: PMC3952453
- DOI: 10.1182/blood-2012-10-451237
The role of chromatin modifiers in normal and malignant hematopoiesis
Abstract
Complex developmental processes such as hematopoiesis require a series of precise and coordinated changes in cellular identity to ensure blood homeostasis. Epigenetic mechanisms help drive changes in gene expression that accompany the transition from hematopoietic stem cells to terminally differentiated blood cells. Genome-wide profiling technologies now provide valuable glimpses of epigenetic changes that occur during normal hematopoiesis, and genetic mouse models developed to investigate the in vivo functions of chromatin-modifying enzymes clearly demonstrate significant roles for these enzymes during embryonic and adult hematopoiesis. Here, we will review the basic science aspects of chromatin modifications and the enzymes that add, remove, and interpret these epigenetic marks. This overview will provide a framework for understanding the roles that these molecules play during normal hematopoiesis. Moreover, many chromatin-modifying enzymes are involved in hematologic malignancies, underscoring the importance of establishing and maintaining appropriate chromatin modification patterns to normal hematology.
Figures

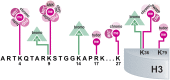
References
-
- O’Connell RM, Baltimore D. MicroRNAs and hematopoietic cell development. Curr Top Dev Biol. 2012;99:145–174. - PubMed
-
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13(1):7–13. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

