The epigenetic landscape of oral squamous cell carcinoma
- PMID: 23287992
- PMCID: PMC3566828
- DOI: 10.1038/bjc.2012.568
The epigenetic landscape of oral squamous cell carcinoma
Abstract
Background: There is relatively little methylation array data available specifically for oral squamous cell carcinoma (OSCC). This study aims to compare the DNA methylome across a large cohort of tumour/normal pairs.
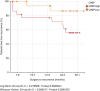
Methods: DNA was extracted from 44 OSCCs and paired normal mucosa. DNA methylation analysis employed the Illumina GoldenGate high-throughput array comprising 1505 CpG loci selected from 807 epigenetically regulated genes. This data was correlated with extracapsular spread (ECS), human papilloma virus (HPV) status, recurrence and 5-year survival.
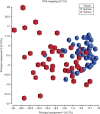
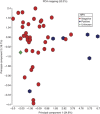
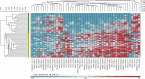
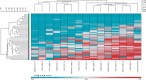
Results: Differential methylation levels of a number of genes distinguished the tumour tissue sample from the matched normal. Putative methylation signatures for ECS and recurrence were identified. The concept of concordant methylation or CpG island methylator phenotype (CIMP) in OSCC is supported by our data, with an association between 'CIMP-high' and worse prognosis. Epigenetic deregulation of NOTCH4 signalling in OSCC was also observed, as part of a possible methylation signature for recurrence, with parallels to recently discovered NOTCH mutations in HNSCC. Differences in methylation in HPV-driven cases were seen, but are less significant than that has been recently proposed in other series.
Conclusion: Although OSCC seems as much an 'epigenetic' as a genetic disease, the translational potential of cancer epigenetics has yet to be fully exploited. This data points to the application of epigenetic biomarkers and targets available to further the development of therapy in OSCC.
Figures
Similar articles
-
Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India.Clin Epigenetics. 2017 Feb 3;9:13. doi: 10.1186/s13148-017-0314-x. eCollection 2017. Clin Epigenetics. 2017. PMID: 28174608 Free PMC article.
-
Global analysis of DNA methylation changes during progression of oral cancer.Oral Oncol. 2013 Nov;49(11):1033-42. doi: 10.1016/j.oraloncology.2013.08.005. Epub 2013 Sep 13. Oral Oncol. 2013. PMID: 24035722
-
Epigenetic deregulation of the anaplastic lymphoma kinase gene modulates mesenchymal characteristics of oral squamous cell carcinomas.Carcinogenesis. 2013 Aug;34(8):1717-27. doi: 10.1093/carcin/bgt112. Epub 2013 Apr 8. Carcinogenesis. 2013. PMID: 23568951 Free PMC article.
-
DNA methylation in oral squamous cell carcinoma: molecular mechanisms and clinical implications.Oral Dis. 2011 Nov;17(8):771-8. doi: 10.1111/j.1601-0825.2011.01833.x. Epub 2011 Jul 22. Oral Dis. 2011. PMID: 21781230 Review.
-
Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype.Histol Histopathol. 2013 May;28(5):585-95. doi: 10.14670/HH-28.585. Epub 2013 Jan 23. Histol Histopathol. 2013. PMID: 23341177 Review.
Cited by
-
METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m6A Methylation in Oral Squamous Cell Carcinoma.Mol Ther. 2020 Oct 7;28(10):2177-2190. doi: 10.1016/j.ymthe.2020.06.024. Epub 2020 Jun 24. Mol Ther. 2020. PMID: 32621798 Free PMC article.
-
Machine Learning-Based Genome-Wide Salivary DNA Methylation Analysis for Identification of Noninvasive Biomarkers in Oral Cancer Diagnosis.Cancers (Basel). 2022 Oct 8;14(19):4935. doi: 10.3390/cancers14194935. Cancers (Basel). 2022. PMID: 36230858 Free PMC article.
-
Anti-Proliferative Effect of Statins Is Mediated by DNMT1 Inhibition and p21 Expression in OSCC Cells.Cancers (Basel). 2020 Jul 28;12(8):2084. doi: 10.3390/cancers12082084. Cancers (Basel). 2020. PMID: 32731382 Free PMC article.
-
Prognostic and therapeutic prediction by screening signature combinations from transcriptome-methylome interactions in oral squamous cell carcinoma.Sci Rep. 2022 Jul 6;12(1):11400. doi: 10.1038/s41598-022-15534-7. Sci Rep. 2022. PMID: 35794182 Free PMC article.
-
Overexpression of suppressor of zest 12 is associated with cervical node metastasis and unfavorable prognosis in tongue squamous cell carcinoma.Cancer Cell Int. 2017 Feb 13;17:26. doi: 10.1186/s12935-017-0395-9. eCollection 2017. Cancer Cell Int. 2017. PMID: 28228691 Free PMC article.
References
-
- Abe M, Ohira M, Kaneda A, Yagi Y, Yamamoto S, Kitano Y, Takato T, Nakagawara A, Ushijima T. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65 (3:828–834. - PubMed
-
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333 (6046:1154–1157. - PMC - PubMed
-
- Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–163. - PubMed
-
- Brock MV, Gou M, Akiyama Y, Muller A, Wu TT, Montgomery E, Deasel M, Germonpre P, Rubinson L, Heitmiller RF, Yang SC, Forastiere AA, Baylin SB, Herman JG. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin Cancer Res. 2003;9 (8:2912–2919. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

