Histone chaperones in nucleosome assembly and human disease
- PMID: 23288364
- PMCID: PMC4004355
- DOI: 10.1038/nsmb.2461
Histone chaperones in nucleosome assembly and human disease
Abstract
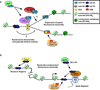
Nucleosome assembly following DNA replication, DNA repair and gene transcription is critical for the maintenance of genome stability and epigenetic information. Nucleosomes are assembled by replication-coupled or replication-independent pathways with the aid of histone chaperone proteins. How these different nucleosome assembly pathways are regulated remains relatively unclear. Recent studies have provided insight into the mechanisms and the roles of histone chaperones in regulating nucleosome assembly. Alterations or mutations in factors involved in nucleosome assembly have also been implicated in cancer and other human diseases. This review highlights the recent progress and outlines future challenges in the field.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

