Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein
- PMID: 23288701
- PMCID: PMC4083568
- DOI: 10.1002/path.4158
Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein
Abstract
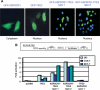
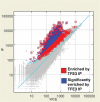
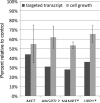
Oncogenic rearrangements of the TFE3 transcription factor gene are found in two distinct human cancers. These include ASPSCR1-TFE3 in all cases of alveolar soft part sarcoma (ASPS) and ASPSCR1-TFE3, PRCC-TFE3, SFPQ-TFE3 and others in a subset of paediatric and adult RCCs. Here we examined the functional properties of the ASPSCR1-TFE3 fusion oncoprotein, defined its target promoters on a genome-wide basis and performed a high-throughput RNA interference screen to identify which of its transcriptional targets contribute to cancer cell proliferation. We first confirmed that ASPSCR1-TFE3 has a predominantly nuclear localization and functions as a stronger transactivator than native TFE3. Genome-wide location analysis performed on the FU-UR-1 cell line, which expresses endogenous ASPSCR1-TFE3, identified 2193 genes bound by ASPSCR1-TFE3. Integration of these data with expression profiles of ASPS tumour samples and inducible cell lines expressing ASPSCR1-TFE3 defined a subset of 332 genes as putative up-regulated direct targets of ASPSCR1-TFE3, including MET (a previously known target gene) and 64 genes as down-regulated targets of ASPSCR1-TFE3. As validation of this approach to identify genuine ASPSCR1-TFE3 target genes, two up-regulated genes bound by ASPSCR1-TFE3, CYP17A1 and UPP1, were shown by multiple lines of evidence to be direct, endogenous targets of transactivation by ASPSCR1-TFE3. As the results indicated that ASPSCR1-TFE3 functions predominantly as a strong transcriptional activator, we hypothesized that a subset of its up-regulated direct targets mediate its oncogenic properties. We therefore chose 130 of these up-regulated direct target genes to study in high-throughput RNAi screens, using FU-UR-1 cells. In addition to MET, we provide evidence that 11 other ASPSCR1-TFE3 target genes contribute to the growth of ASPSCR1-TFE3-positive cells. Our data suggest new therapeutic possibilities for cancers driven by TFE3 fusions. More generally, this work establishes a combined integrated genomics/functional genomics strategy to dissect the biology of oncogenic, chimeric transcription factors.
Copyright © 2013 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Figures




References
-
- Ladanyi M, Lui MY, Antonescu CR, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. - PubMed
-
- Kayton ML, Meyers P, Wexler LH, et al. Clinical presentation, treatment, and outcome of alveolar soft part sarcoma in children, adolescents, and young adults. J Pediatr Surg. 2006;41:187–193. - PubMed
-
- Argani P, Olgac S, Tickoo SK, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–1160. - PubMed
-
- Ishiguro M, Iwasaki H, Ohjimi Y, et al. Establishment and characterization of a renal cell carcinoma cell line (FU-UR-1) with the reciprocal ASPL–TFE3 fusion transcript. Oncol Rep. 2004;11:1169–1175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

