Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills
- PMID: 23288898
- PMCID: PMC3557025
- DOI: 10.1073/pnas.1213892110
Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills
Abstract
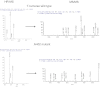
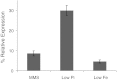
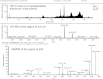
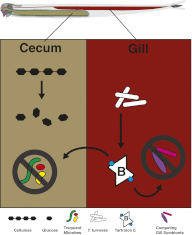
Shipworms are marine wood-boring bivalve mollusks (family Teredinidae) that harbor a community of closely related Gammaproteobacteria as intracellular endosymbionts in their gills. These symbionts have been proposed to assist the shipworm host in cellulose digestion and have been shown to play a role in nitrogen fixation. The genome of one strain of Teredinibacter turnerae, the first shipworm symbiont to be cultivated, was sequenced, revealing potential as a rich source of polyketides and nonribosomal peptides. Bioassay-guided fractionation led to the isolation and identification of two macrodioloide polyketides belonging to the tartrolon class. Both compounds were found to possess antibacterial properties, and the major compound was found to inhibit other shipworm symbiont strains and various pathogenic bacteria. The gene cluster responsible for the synthesis of these compounds was identified and characterized, and the ketosynthase domains were analyzed phylogenetically. Reverse-transcription PCR in addition to liquid chromatography and high-resolution mass spectrometry and tandem mass spectrometry revealed the transcription of these genes and the presence of the compounds in the shipworm, suggesting that the gene cluster is expressed in vivo and that the compounds may fulfill a specific function for the shipworm host. This study reports tartrolon polyketides from a shipworm symbiont and unveils the biosynthetic gene cluster of a member of this class of compounds, which might reveal the mechanism by which these bioactive metabolites are biosynthesized.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Distel DL. The biology of marine wood boring bivalves and their bacterial endosymbionts. In: Goodell B, Nicholas D, Schultz T, editors. Wood Deterioration and Preservation. Washington, DC: American Chemical Society; 2003. pp. 253–271.
-
- Ohkuma M. Termite symbiotic systems: Efficient bio-recycling of lignocellulose. Appl Microbiol Biotechnol. 2003;61(1):1–9. - PubMed
-
- Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. - PubMed
-
- Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J. Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae) Int J Syst Evol Microbiol. 2002;52(Pt 6):2261–2269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

