Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli
- PMID: 23288927
- PMCID: PMC3571447
- DOI: 10.1093/infdis/jis933
Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli
Abstract
Background: Fluoroquinolone-resistant Escherichia coli are increasingly prevalent. Their clonal origins--potentially critical for control efforts--remain undefined.
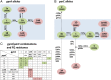
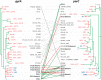
Methods: Antimicrobial resistance profiles and fine clonal structure were determined for 236 diverse-source historical (1967-2009) E. coli isolates representing sequence type ST131 and 853 recent (2010-2011) consecutive E. coli isolates from 5 clinical laboratories in Seattle, Washington, and Minneapolis, Minnesota. Clonal structure was resolved based on fimH sequence (fimbrial adhesin gene: H subclone assignments), multilocus sequence typing, gyrA and parC sequence (fluoroquinolone resistance-determining loci), and pulsed-field gel electrophoresis.
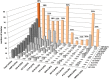
Results: Of the recent fluoroquinolone-resistant clinical isolates, 52% represented a single ST131 subclonal lineage, H30, which expanded abruptly after 2000. This subclone had a unique and conserved gyrA/parC allele combination, supporting its tight clonality. Unlike other ST131 subclones, H30 was significantly associated with fluoroquinolone resistance and was the most prevalent subclone among current E. coli clinical isolates, overall (10.4%) and within every resistance category (11%-52%).
Conclusions: Most current fluoroquinolone-resistant E. coli clinical isolates, and the largest share of multidrug-resistant isolates, represent a highly clonal subgroup that likely originated from a single rapidly expanded and disseminated ST131 strain. Focused attention to this strain will be required to control the fluoroquinolone and multidrug-resistant E. coli epidemic.
Figures


Comment in
-
Response to Giufre et al.J Infect Dis. 2014 Feb 15;209(4):630-1. doi: 10.1093/infdis/jit582. Epub 2013 Nov 7. J Infect Dis. 2014. PMID: 24203777 Free PMC article. No abstract available.
-
Predominance of the fimH30 subclone among multidrug-resistant Escherichia coli strains belonging to sequence type 131 in Italy.J Infect Dis. 2014 Feb 15;209(4):629-30. doi: 10.1093/infdis/jit583. Epub 2013 Nov 7. J Infect Dis. 2014. PMID: 24203778 No abstract available.
References
-
- Gilbert DN, Moellering RCJ, Eliopoulos GM, Chambers HF, Saag MS. The Sanford Guide to Antimicrobial Therapy 2010. 40th ed. Vienna, VA: Antimicrobial Therapy, Inc; 2010. pp. 4–64.
-
- Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect. 2003;5:449–56. - PubMed
-
- Tufan Z, Bulut C, Yazan T, et al. A life-threatening Escherichia coli meningitis after prostate biopsy. Urol J. 2011;8:69–71. - PubMed
-
- Gupta K, Hooton TM, Naber KG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52:e103–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

