A simple method for predicting transmembrane proteins based on wavelet transform
- PMID: 23289014
- PMCID: PMC3535531
- DOI: 10.7150/ijbs.5371
A simple method for predicting transmembrane proteins based on wavelet transform
Abstract
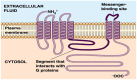
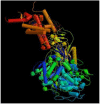
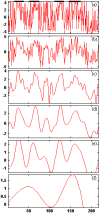
The increasing protein sequences from the genome project require theoretical methods to predict transmembrane helical segments (TMHs). So far, several prediction methods have been reported, but there are some deficiencies in prediction accuracy and adaptability in these methods. In this paper, a method based on discrete wavelet transform (DWT) has been developed to predict the number and location of TMHs in membrane proteins. PDB coded as 1KQG is chosen as an example to describe the prediction process by this method. 80 proteins with known 3D structure from Mptopo database are chosen at random as data sets (including 325 TMHs) and 80 sequences are divided into 13 groups according to their function and type. TMHs prediction is carried out for each group of membrane protein sequences and obtain satisfactory result. To verify the feasibility of this method, 80 membrane protein sequences are treated as test sets, 308 TMHs can be predicted and the prediction accuracy is 96.3%. Compared with the main prediction results of seven popular prediction methods, the obtained results indicate that the proposed method in this paper has higher prediction accuracy.
Keywords: Discrete wavelet transform; Hydrophobicity.; Membrane protein; Transmembrane helical segments.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

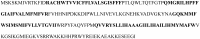
References
-
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. - PubMed
-
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nature Biotechnology. 2007;25:1119–1126. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

