Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer
- PMID: 23292282
- PMCID: PMC5417325
- DOI: 10.1210/me.2012-1265
Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer
Abstract
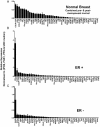
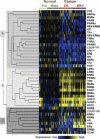
To identify biologically relevant groupings or clusters of nuclear receptors (NR) that are associated with breast neoplasia, with potentially diagnostic, discriminant or prognostic value, we quantitated mRNA expression levels of all 48 members of the human NR superfamily by TaqMan low-density array analysis in 116 curated breast tissue samples, including pre- and postmenopausal normal breast and both ERα(+) and ERα(-) tumor tissue. In addition, we have determined NR levels in independent cohorts of tamoxifen-treated ERα(+) and ERα(-) tissue samples. There were differences in relative NR mRNA expression between neoplastic and normal breast, and between ER(+) and ER(-) tumors. First, there is overexpression of the NUR77 subgroup and EAR2 in neoplastic breast. Second, we identify a signature of five NR (ERα, EAR2, NUR77, TRα, and RARγ) that classifies breast samples with more than 97% cross-validated accuracy into normal or cancer classes. Third, we find a novel negative association between five NR (TRβ, NUR77, RORγ, COUP-TFII, and LRH1) and histological grade. Finally, four NR (COUP-TFII, TRβ, PPARγ, and MR) are significant predictors of metastasis-free survival in tamoxifen-treated breast cancers, independent of ER expression. The present study highlights the discriminant and prognostic value of NR in breast cancer; identifies novel, clinically relevant, NR signatures; and highlights NR signaling pathways with potential roles in breast cancer pathophysiology and as new therapeutic targets.
Figures





References
-
- Cuzick J , Sestak I , Baum M , Buzdar A , Howell A , Dowsett M , Forbes JF. 2010. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol 11:1135–1141 - PubMed
-
- Arteaga CL , Sliwkowski MX , Osborne CK , Perez EA , Puglisi F , Gianni L. 2012. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol 9:16–32 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

