Novel twin streptolysin S-like peptides encoded in the sag operon homologue of beta-hemolytic Streptococcus anginosus
- PMID: 23292771
- PMCID: PMC3571330
- DOI: 10.1128/JB.01344-12
Novel twin streptolysin S-like peptides encoded in the sag operon homologue of beta-hemolytic Streptococcus anginosus
Abstract
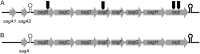
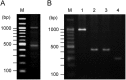
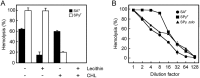
Streptococcus anginosus is a member of the anginosus group streptococci, which form part of the normal human oral flora. In contrast to the pyogenic group streptococci, our knowledge of the virulence factors of the anginosus group streptococci, including S. anginosus, is not sufficient to allow a clear understanding of the basis of their pathogenicity. Generally, hemolysins are thought to be important virulence factors in streptococcal infections. In the present study, a sag operon homologue was shown to be responsible for beta-hemolysis in S. anginosus strains by random gene knockout. Interestingly, contrary to pyogenic group streptococci, beta-hemolytic S. anginosus was shown to have two tandem sagA homologues, encoding streptolysin S (SLS)-like peptides, in the sag operon homologue. Gene deletion and complementation experiments revealed that both genes were functional, and these SLS-like peptides were essential for beta-hemolysis in beta-hemolytic S. anginosus. Furthermore, the amino acid sequence of these SLS-like peptides differed from that of the typical SLS of S. pyogenes, especially in their propeptide domain, and an amino acid residue indicated to be important for the cytolytic activity of SLS in S. pyogenes was deleted in both S. anginosus homologues. These data suggest that SLS-like peptides encoded by two sagA homologues in beta-hemolytic S. anginosus may be potential virulence factors with a different structure essential for hemolytic activity and/or the maturation process compared to the typical SLS present in pyogenic group streptococci.
Figures






References
-
- Whiley RA, Beighton D. 1991. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as distinct species. Int. J. Syst. Bacteriol. 41:1–5 - PubMed
-
- Whiley RA, Hall LMC, Hardie JM, Beighton D. 1999. A study of small-colony, β-hemolytic, Lancefield group C streptococci within the anginosus group: description of Streptococcus constellatus subsp. pharynges subsp. nov., associated with the human throat and pharyngitis. Int. J. Syst. Bacteriol. 49:1443–1449 - PubMed
-
- Clarridge JE, III, Attorri S, Musher DM, Hebert J, Dunber S. 2001. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (“Streptococcus milleri group”) are of different clinical importance and are not equally associated with abscess. Clin. Infect. Dis. 32:1511–1515 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

