DNA looping in prokaryotes: experimental and theoretical approaches
- PMID: 23292776
- PMCID: PMC3591992
- DOI: 10.1128/JB.02038-12
DNA looping in prokaryotes: experimental and theoretical approaches
Abstract
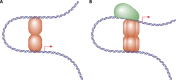
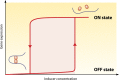
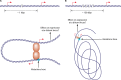
Transcriptional regulation is at the heart of biological functions such as adaptation to a changing environment or to new carbon sources. One of the mechanisms which has been found to modulate transcription, either positively (activation) or negatively (repression), involves the formation of DNA loops. A DNA loop occurs when a protein or a complex of proteins simultaneously binds to two different sites on DNA with looping out of the intervening DNA. This simple mechanism is central to the regulation of several operons in the genome of the bacterium Escherichia coli, like the lac operon, one of the paradigms of genetic regulation. The aim of this review is to gather and discuss concepts and ideas from experimental biology and theoretical physics concerning DNA looping in genetic regulation. We first describe experimental techniques designed to show the formation of a DNA loop. We then present the benefits that can or could be derived from a mechanism involving DNA looping. Some of these are already experimentally proven, but others are theoretical predictions and merit experimental investigation. Then, we try to identify other genetic systems that could be regulated by a DNA looping mechanism in the genome of Escherichia coli. We found many operons that, according to our set of criteria, have a good chance to be regulated with a DNA loop. Finally, we discuss the proposition recently made by both biologists and physicists that this mechanism could also act at the genomic scale and play a crucial role in the spatial organization of genomes.
Figures
References
-
- Dame RT, Kalmykowa OJ, Grainger DC. 2011. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in Gram negative bacteria. PLoS Genet. 7:e1002123 doi:10.1371/journal.pgen.1002123 - DOI - PMC - PubMed
-
- Pettijohn D. 1996. The nucleoid, p 158–166 In Neidhardt F. C., Curtiss R., III, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M, Umbarger H. E. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC
-
- Kavenoff R, Bowen BC. 1976. Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma 59:89–101 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

