Genetic heterogeneity of diffuse large B-cell lymphoma
- PMID: 23292937
- PMCID: PMC3557051
- DOI: 10.1073/pnas.1205299110
Genetic heterogeneity of diffuse large B-cell lymphoma
Abstract
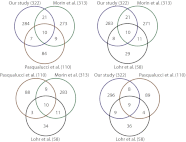
Diffuse large B-cell lymphoma (DLBCL) is the most common form of lymphoma in adults. The disease exhibits a striking heterogeneity in gene expression profiles and clinical outcomes, but its genetic causes remain to be fully defined. Through whole genome and exome sequencing, we characterized the genetic diversity of DLBCL. In all, we sequenced 73 DLBCL primary tumors (34 with matched normal DNA). Separately, we sequenced the exomes of 21 DLBCL cell lines. We identified 322 DLBCL cancer genes that were recurrently mutated in primary DLBCLs. We identified recurrent mutations implicating a number of known and not previously identified genes and pathways in DLBCL including those related to chromatin modification (ARID1A and MEF2B), NF-κB (CARD11 and TNFAIP3), PI3 kinase (PIK3CD, PIK3R1, and MTOR), B-cell lineage (IRF8, POU2F2, and GNA13), and WNT signaling (WIF1). We also experimentally validated a mutation in PIK3CD, a gene not previously implicated in lymphomas. The patterns of mutation demonstrated a classic long tail distribution with substantial variation of mutated genes from patient to patient and also between published studies. Thus, our study reveals the tremendous genetic heterogeneity that underlies lymphomas and highlights the need for personalized medicine approaches to treating these patients.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. - PubMed
-
- Monti S, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105(5):1851–1861. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

